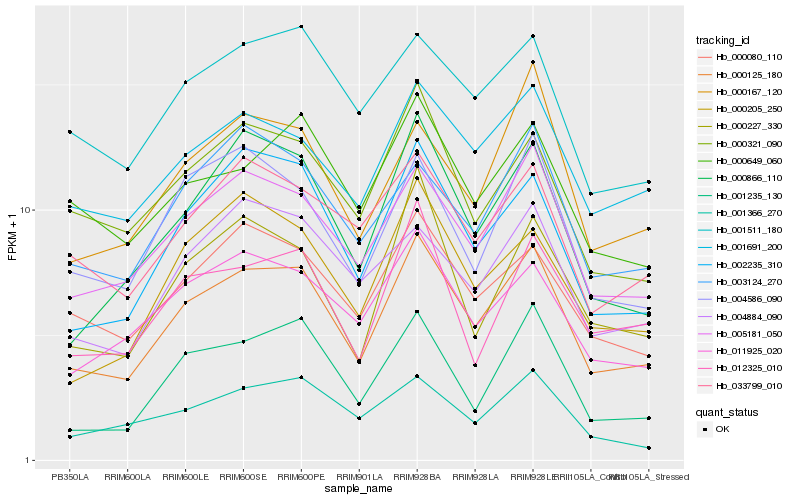
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000125_180 |
0.0 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 2 |
Hb_005181_050 |
0.070391414 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 3 |
Hb_002235_310 |
0.0763647863 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 4 |
Hb_000866_110 |
0.0866862607 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 5 |
Hb_011925_020 |
0.089846467 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 6 |
Hb_001691_200 |
0.0902294879 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 7 |
Hb_000080_110 |
0.0902981181 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000321_090 |
0.0945497607 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 9 |
Hb_000167_120 |
0.0954605239 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 10 |
Hb_004586_090 |
0.0956911641 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000205_250 |
0.0978362462 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 12 |
Hb_033799_010 |
0.0980179002 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 13 |
Hb_012325_010 |
0.0980633476 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 14 |
Hb_001235_130 |
0.0982345067 |
- |
- |
- |
| 15 |
Hb_000227_330 |
0.0984357548 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 16 |
Hb_001511_180 |
0.0986569468 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 17 |
Hb_004884_090 |
0.0998750128 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 18 |
Hb_003124_270 |
0.1005124273 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 19 |
Hb_000649_060 |
0.1012081917 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 20 |
Hb_001366_270 |
0.1025105816 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |