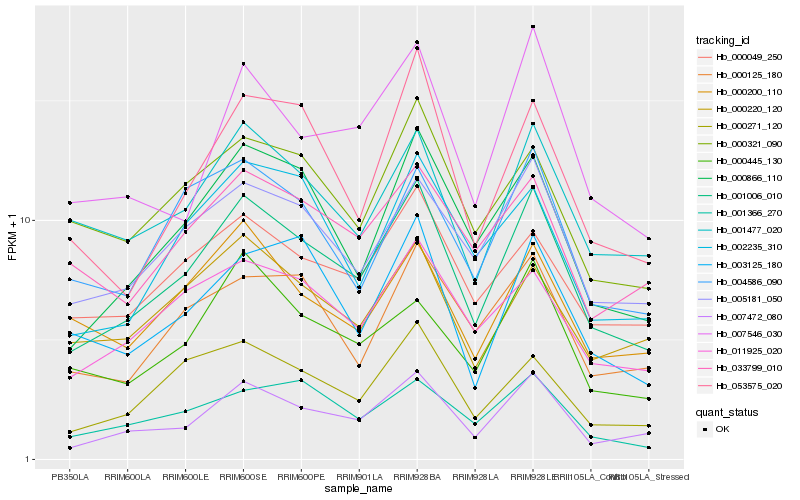
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001006_010 |
0.0 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 2 |
Hb_007472_080 |
0.0676842536 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 3 |
Hb_000866_110 |
0.0882424942 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005181_050 |
0.0923522818 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 5 |
Hb_000271_120 |
0.0965438452 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 6 |
Hb_011925_020 |
0.0972717492 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 7 |
Hb_000049_250 |
0.1014901094 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001477_020 |
0.1029355673 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 9 |
Hb_001366_270 |
0.1050410603 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 10 |
Hb_007546_030 |
0.1062077594 |
- |
- |
transcription cofactor, putative [Ricinus communis] |
| 11 |
Hb_000125_180 |
0.1081173282 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 12 |
Hb_002235_310 |
0.1102327255 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 13 |
Hb_053575_020 |
0.1103329182 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 14 |
Hb_000445_130 |
0.1103665754 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 15 |
Hb_000321_090 |
0.111386809 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 16 |
Hb_000220_120 |
0.1116553945 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004586_090 |
0.1118429406 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003125_180 |
0.1142060409 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 19 |
Hb_000200_110 |
0.1142136292 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 20 |
Hb_033799_010 |
0.1147614916 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |