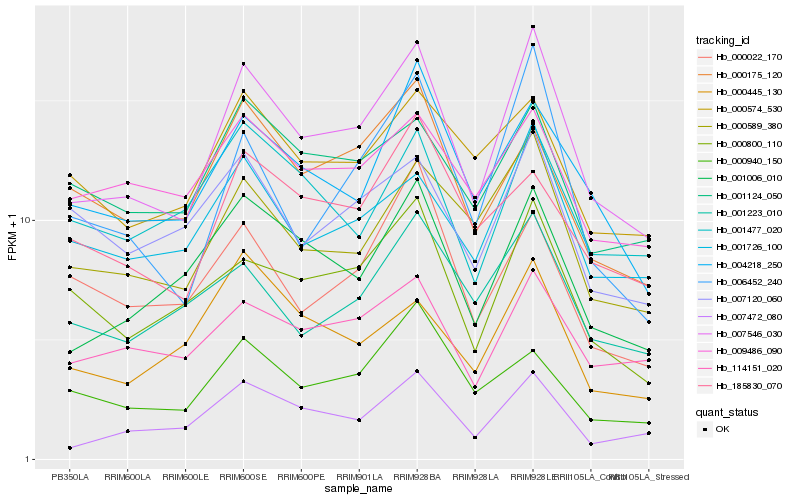
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007546_030 |
0.0 |
- |
- |
transcription cofactor, putative [Ricinus communis] |
| 2 |
Hb_007472_080 |
0.1040516171 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 3 |
Hb_001006_010 |
0.1062077594 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 4 |
Hb_000589_380 |
0.109703034 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 5 |
Hb_001726_100 |
0.1156538648 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000800_110 |
0.1175168762 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_004218_250 |
0.120414391 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 8 |
Hb_006452_240 |
0.1210880038 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 15a isoform X1 [Jatropha curcas] |
| 9 |
Hb_114151_020 |
0.1235826161 |
- |
- |
glucan water dikinase 2 [Manihot esculenta] |
| 10 |
Hb_000175_120 |
0.1318899464 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 11 |
Hb_001223_010 |
0.1319631719 |
- |
- |
PREDICTED: FGGY carbohydrate kinase domain-containing protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_001124_050 |
0.1321014802 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 13 |
Hb_007120_060 |
0.1347483654 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 14 |
Hb_001477_020 |
0.1367709189 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000445_130 |
0.1371134247 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 16 |
Hb_185830_070 |
0.1375563371 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000940_150 |
0.1379573209 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 18 |
Hb_000574_530 |
0.1382034796 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 19 |
Hb_000022_170 |
0.1421282397 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 20 |
Hb_009486_090 |
0.1422419728 |
- |
- |
RNA binding protein, putative [Ricinus communis] |