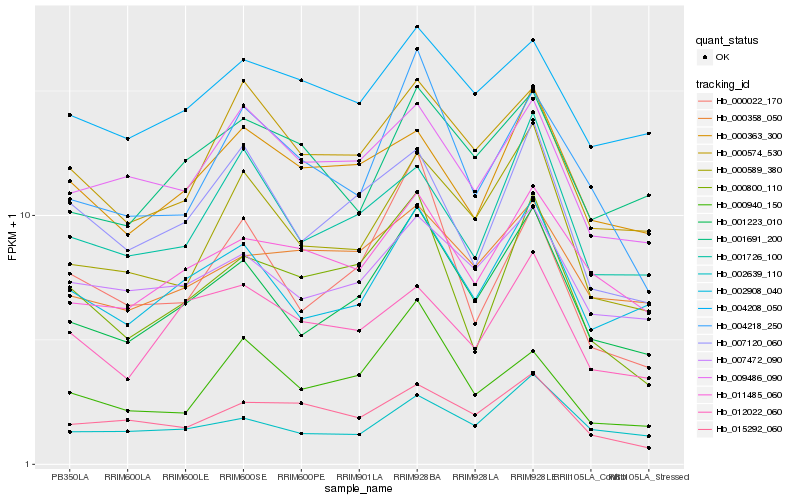
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001223_010 |
0.0 |
- |
- |
PREDICTED: FGGY carbohydrate kinase domain-containing protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_002908_040 |
0.0706777597 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 3 |
Hb_000589_380 |
0.0899241184 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 4 |
Hb_007472_090 |
0.0950063849 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_002639_110 |
0.096538013 |
- |
- |
hypothetical protein L484_011673 [Morus notabilis] |
| 6 |
Hb_011485_060 |
0.1043993225 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 7 |
Hb_001726_100 |
0.1076738615 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007120_060 |
0.1117521034 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 9 |
Hb_000574_530 |
0.1119665149 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 10 |
Hb_004218_250 |
0.1154270936 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 11 |
Hb_000800_110 |
0.1172235605 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000022_170 |
0.1176679398 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 13 |
Hb_009486_090 |
0.1196189705 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000358_050 |
0.120409512 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 15 |
Hb_001691_200 |
0.1207700221 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 16 |
Hb_000363_300 |
0.1222949318 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_012022_060 |
0.123607781 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004208_050 |
0.1247450986 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 19 |
Hb_015292_060 |
0.1270598447 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 20 |
Hb_000940_150 |
0.1271045588 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |