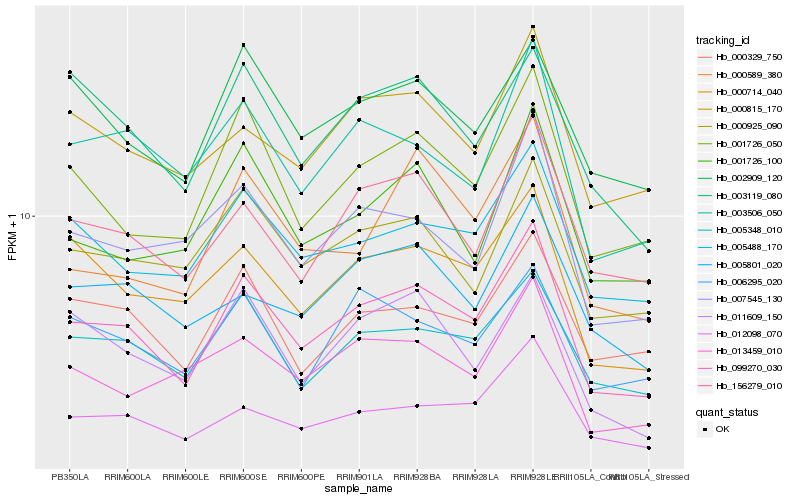
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_099270_030 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001726_050 |
0.0784015515 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 3 |
Hb_005348_010 |
0.0828804722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012098_070 |
0.0864097906 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 5 |
Hb_156279_010 |
0.0871187437 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005488_170 |
0.0871609492 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 7 |
Hb_000714_040 |
0.0880456957 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003506_050 |
0.0904438116 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000925_090 |
0.0940247718 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000329_750 |
0.0949691416 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 11 |
Hb_007545_130 |
0.0951589875 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000815_170 |
0.0959316949 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 13 |
Hb_013459_010 |
0.0965288349 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 14 |
Hb_001726_100 |
0.0990478188 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005801_020 |
0.1019531125 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 16 |
Hb_002909_120 |
0.102350226 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 17 |
Hb_000589_380 |
0.1025962125 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 18 |
Hb_003119_080 |
0.1030251416 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 19 |
Hb_006295_020 |
0.1034178523 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR isoform X1 [Jatropha curcas] |
| 20 |
Hb_011609_150 |
0.1039139015 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |