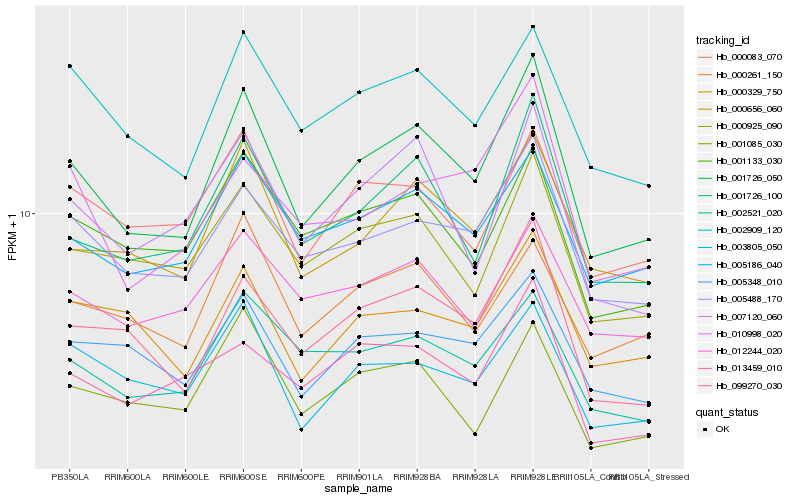
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001726_050 |
0.0 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_001726_100 |
0.068549002 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000656_060 |
0.074417581 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_005488_170 |
0.0759422517 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 5 |
Hb_099270_030 |
0.0784015515 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005186_040 |
0.0826758506 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002909_120 |
0.085928718 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 8 |
Hb_005348_010 |
0.0880861614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002521_020 |
0.0885086758 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000925_090 |
0.088931131 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007120_060 |
0.0907654102 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 12 |
Hb_001133_030 |
0.0919989523 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 13 |
Hb_003805_050 |
0.0931241886 |
- |
- |
- |
| 14 |
Hb_000329_750 |
0.0948523116 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 15 |
Hb_012244_020 |
0.0953717663 |
- |
- |
calpain, putative [Ricinus communis] |
| 16 |
Hb_000083_070 |
0.0954070908 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000261_150 |
0.0954770472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010998_020 |
0.0991971391 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 19 |
Hb_013459_010 |
0.101120694 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 20 |
Hb_001085_030 |
0.102098698 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |