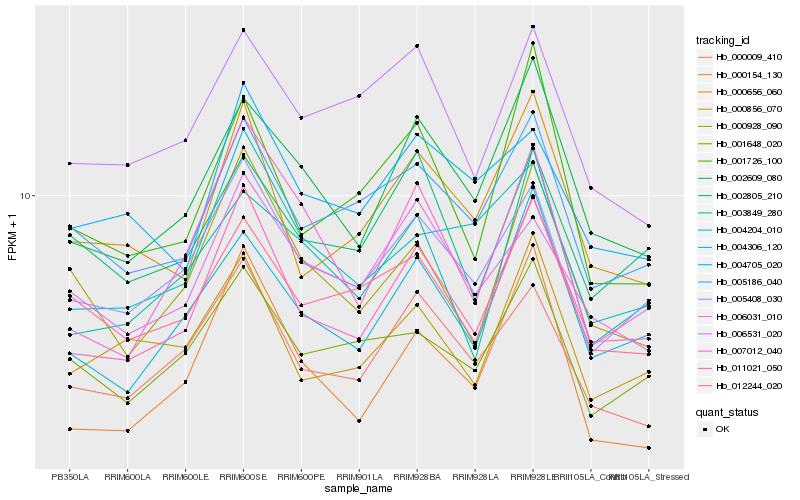
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011021_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004306_120 |
0.0629295699 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_005408_030 |
0.0638497963 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 4 |
Hb_002609_080 |
0.0645964104 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000009_410 |
0.0710450394 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 6 |
Hb_000656_060 |
0.0713253238 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_005186_040 |
0.0763666646 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000856_070 |
0.0791786515 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 9 |
Hb_000928_090 |
0.0800998777 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 10 |
Hb_004204_010 |
0.0823838113 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 11 |
Hb_012244_020 |
0.0848304642 |
- |
- |
calpain, putative [Ricinus communis] |
| 12 |
Hb_006531_020 |
0.0850211625 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 13 |
Hb_007012_040 |
0.086296563 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 14 |
Hb_000154_130 |
0.0863656799 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 15 |
Hb_001648_020 |
0.0867777065 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 16 |
Hb_003849_280 |
0.0882241677 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 17 |
Hb_002805_210 |
0.0897618632 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 18 |
Hb_001726_100 |
0.0906992995 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004705_020 |
0.0910506662 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 20 |
Hb_006031_010 |
0.092576413 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |