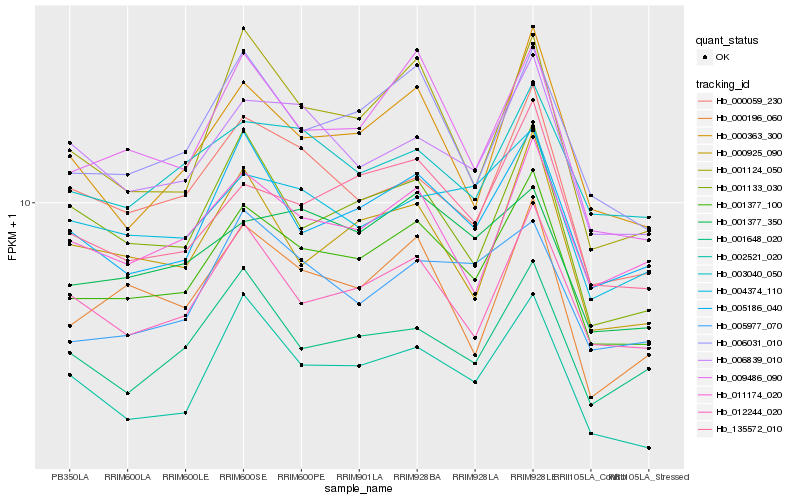
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 2 |
Hb_009486_090 |
0.0549795896 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_006031_010 |
0.0561331211 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 4 |
Hb_135572_010 |
0.0574073783 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 5 |
Hb_005186_040 |
0.0602299004 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 6 |
Hb_012244_020 |
0.0610528165 |
- |
- |
calpain, putative [Ricinus communis] |
| 7 |
Hb_000059_230 |
0.0641898053 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 8 |
Hb_002521_020 |
0.0644840463 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000363_300 |
0.0648685277 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006839_010 |
0.0658534324 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 11 |
Hb_011174_020 |
0.0695298104 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 12 |
Hb_000196_060 |
0.0695688047 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 13 |
Hb_000925_090 |
0.0702738306 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001133_030 |
0.0705164388 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 15 |
Hb_001377_350 |
0.070907413 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 16 |
Hb_001124_050 |
0.0723117679 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 17 |
Hb_003040_050 |
0.0731701575 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 18 |
Hb_004374_110 |
0.0733988968 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 19 |
Hb_005977_070 |
0.0744847386 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001648_020 |
0.0746802912 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |