| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
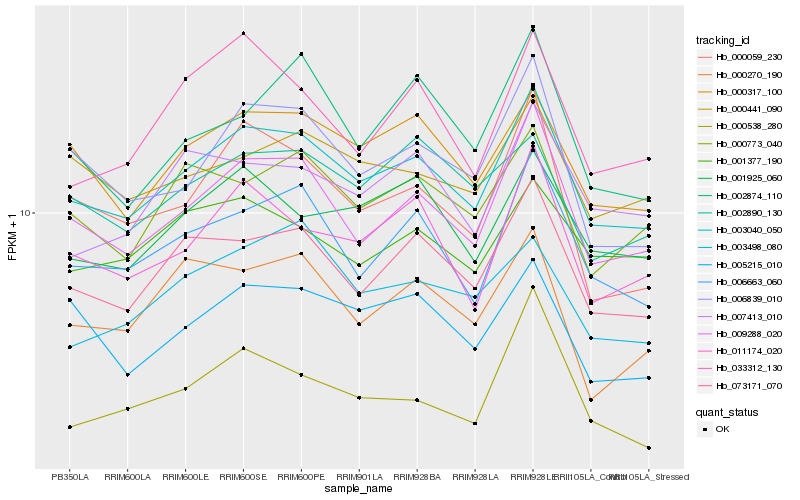
Hb_003040_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 2 |
Hb_009288_020 |
0.0377808639 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 3 |
Hb_073171_070 |
0.042135891 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 4 |
Hb_006663_060 |
0.0495268988 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 5 |
Hb_000317_100 |
0.0499223671 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 6 |
Hb_011174_020 |
0.0563025109 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 7 |
Hb_001925_060 |
0.0565103231 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 8 |
Hb_006839_010 |
0.0594876324 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 9 |
Hb_000773_040 |
0.0597121993 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005215_010 |
0.0598627889 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000441_090 |
0.0623807606 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 12 |
Hb_002890_130 |
0.0635335498 |
- |
- |
tip120, putative [Ricinus communis] |
| 13 |
Hb_003498_080 |
0.0643023601 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 14 |
Hb_001377_190 |
0.0650432705 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007413_010 |
0.0655956201 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 16 |
Hb_000538_280 |
0.0657767038 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 17 |
Hb_000059_230 |
0.066729244 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 18 |
Hb_000270_190 |
0.0669957251 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 19 |
Hb_002874_110 |
0.0681649339 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 20 |
Hb_033312_130 |
0.0681856161 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |