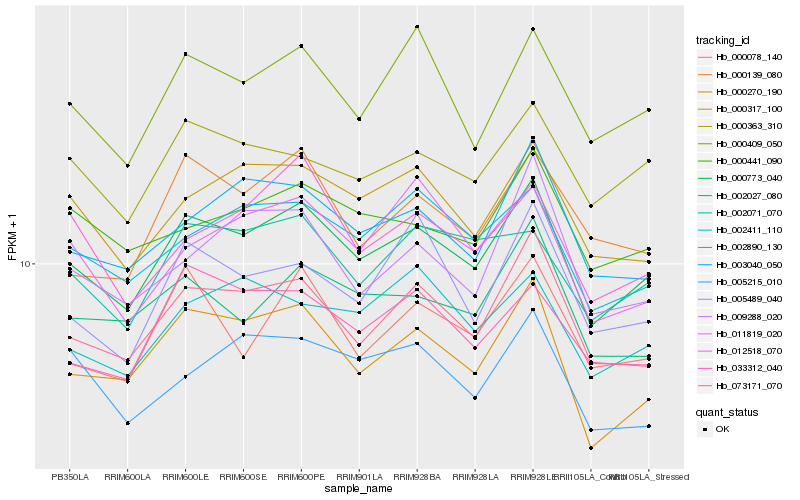
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000773_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 2 |
Hb_073171_070 |
0.0468721135 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 3 |
Hb_000270_190 |
0.0490262684 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 4 |
Hb_000317_100 |
0.0580389721 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 5 |
Hb_000409_050 |
0.0581157696 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 6 |
Hb_002890_130 |
0.058507753 |
- |
- |
tip120, putative [Ricinus communis] |
| 7 |
Hb_003040_050 |
0.0597121993 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 8 |
Hb_000363_310 |
0.0608114348 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 9 |
Hb_002027_080 |
0.0625340464 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 10 |
Hb_002071_070 |
0.062571646 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_033312_040 |
0.0637261502 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000441_090 |
0.0645615622 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 13 |
Hb_002411_110 |
0.066870207 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 14 |
Hb_012518_070 |
0.0669001614 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000078_140 |
0.0672203855 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 16 |
Hb_000139_080 |
0.0676148088 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_011819_020 |
0.0699565111 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005489_040 |
0.0705735569 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 19 |
Hb_005215_010 |
0.0706647124 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_009288_020 |
0.0708635129 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |