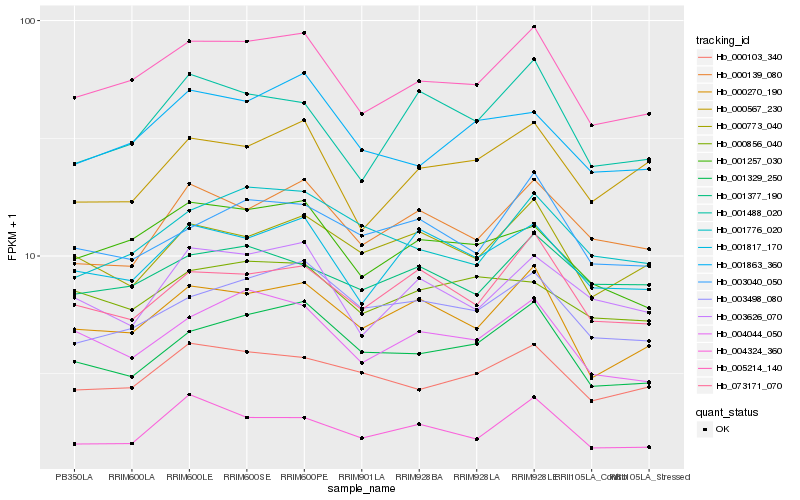
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005214_140 |
0.0 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 2 |
Hb_000270_190 |
0.0573189908 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 3 |
Hb_001257_030 |
0.0605819109 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek5 [Jatropha curcas] |
| 4 |
Hb_004044_050 |
0.06433335 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 5 |
Hb_001817_170 |
0.0651157149 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 6 |
Hb_001329_250 |
0.0655524205 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000103_340 |
0.0684299974 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 8 |
Hb_003498_080 |
0.0692645518 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_073171_070 |
0.0720239063 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 10 |
Hb_001863_360 |
0.0722203165 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 11 |
Hb_003626_070 |
0.0753252309 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 12 |
Hb_000567_230 |
0.0764415923 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 13 |
Hb_004324_360 |
0.0764628102 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000139_080 |
0.0765794227 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001488_020 |
0.0771152825 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 16 |
Hb_000773_040 |
0.077705078 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003040_050 |
0.0780608807 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 18 |
Hb_001377_190 |
0.0794361284 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000856_040 |
0.0800306511 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 20 |
Hb_001776_020 |
0.0813740089 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |