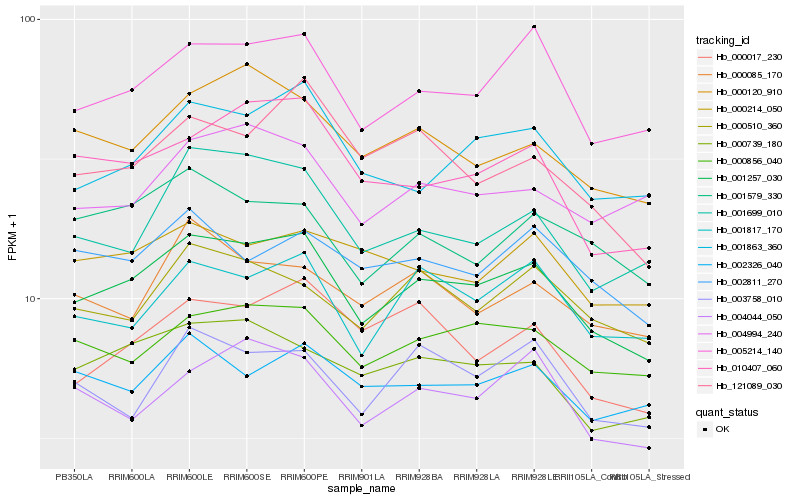
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001257_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek5 [Jatropha curcas] |
| 2 |
Hb_005214_140 |
0.0605819109 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 3 |
Hb_000739_180 |
0.0697757336 |
- |
- |
PREDICTED: uncharacterized protein LOC105643110 [Jatropha curcas] |
| 4 |
Hb_001863_360 |
0.0708057677 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 5 |
Hb_000856_040 |
0.0711076145 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 6 |
Hb_001817_170 |
0.0734782474 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 7 |
Hb_010407_060 |
0.0737292515 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 8 |
Hb_000120_910 |
0.0759604947 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 9 |
Hb_004044_050 |
0.0771481014 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 10 |
Hb_001699_010 |
0.0775337383 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 11 |
Hb_000510_360 |
0.0782300739 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 12 |
Hb_001579_330 |
0.0798661005 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_121089_030 |
0.0800866683 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 14 |
Hb_000017_230 |
0.0819848065 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 15 |
Hb_003758_010 |
0.0821012535 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002811_270 |
0.0829216529 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 17 |
Hb_000085_170 |
0.083143935 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002326_040 |
0.0832479047 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 19 |
Hb_004994_240 |
0.0840911843 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 20 |
Hb_000214_050 |
0.0845648243 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |