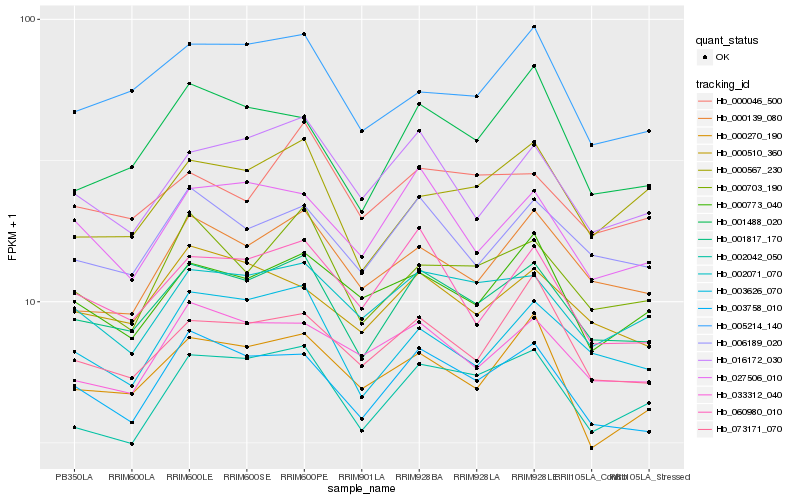
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001817_170 |
0.0 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 2 |
Hb_003758_010 |
0.0468744969 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006189_020 |
0.0538738375 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 4 |
Hb_002042_050 |
0.0583150063 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003626_070 |
0.0589868302 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 6 |
Hb_002071_070 |
0.0627296014 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000139_080 |
0.0634234489 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000510_360 |
0.0639301342 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 9 |
Hb_000703_190 |
0.0648982786 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005214_140 |
0.0651157149 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 11 |
Hb_073171_070 |
0.0662006688 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 12 |
Hb_001488_020 |
0.0671797637 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 13 |
Hb_033312_040 |
0.0679912576 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_060980_010 |
0.0682153471 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 15 |
Hb_027506_010 |
0.0692889507 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 16 |
Hb_016172_030 |
0.0693858465 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 17 |
Hb_000270_190 |
0.0719303622 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 18 |
Hb_000773_040 |
0.0727328606 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000567_230 |
0.0727422075 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 20 |
Hb_000046_500 |
0.0730316748 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |