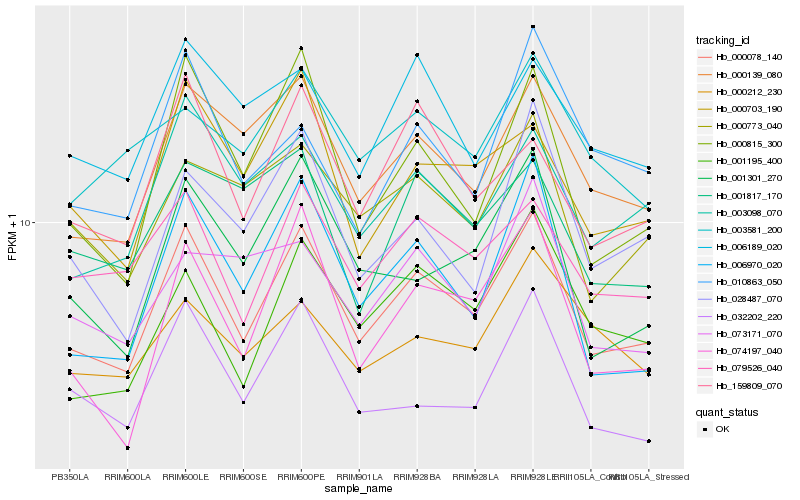
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000078_140 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 2 |
Hb_000139_080 |
0.0554180329 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_079526_040 |
0.0589348608 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 4 |
Hb_074197_040 |
0.0600618403 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 5 |
Hb_032202_220 |
0.0602803397 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_003098_070 |
0.0603273167 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 7 |
Hb_001195_400 |
0.0619800791 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 8 |
Hb_159809_070 |
0.0623997762 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 9 |
Hb_000703_190 |
0.0635932387 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000815_300 |
0.0653859755 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 11 |
Hb_006970_020 |
0.0660578456 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 12 |
Hb_000773_040 |
0.0672203855 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 13 |
Hb_006189_020 |
0.0679768061 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 14 |
Hb_001301_270 |
0.0691106643 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 15 |
Hb_028487_070 |
0.0701417056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003581_200 |
0.0714472339 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 17 |
Hb_010863_050 |
0.074614843 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 18 |
Hb_000212_230 |
0.0751715942 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 19 |
Hb_073171_070 |
0.0754649963 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 20 |
Hb_001817_170 |
0.0758338891 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |