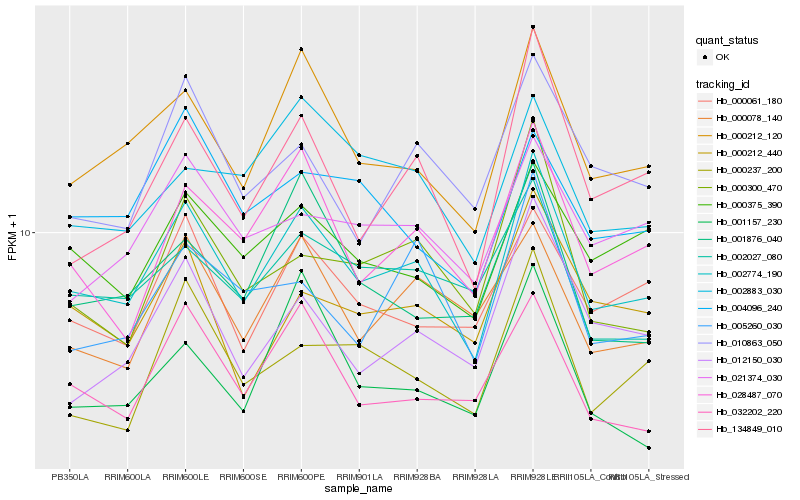
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002774_190 |
0.0 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 2 |
Hb_000212_120 |
0.0517214934 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 3 |
Hb_028487_070 |
0.0683944436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_032202_220 |
0.0710356346 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_134849_010 |
0.0714306125 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 6 |
Hb_000375_390 |
0.0720150862 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 7 |
Hb_000061_180 |
0.0743509938 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 8 |
Hb_000078_140 |
0.0766391246 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 9 |
Hb_000237_200 |
0.0766852527 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 10 |
Hb_002027_080 |
0.0769100715 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 11 |
Hb_021374_030 |
0.0782672231 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 12 |
Hb_000300_470 |
0.0789181929 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 13 |
Hb_002883_030 |
0.0790780296 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 14 |
Hb_012150_030 |
0.0797442399 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001876_040 |
0.0805934366 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 16 |
Hb_001157_230 |
0.0807473971 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 17 |
Hb_010863_050 |
0.0810132352 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 18 |
Hb_005260_030 |
0.0811200856 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 19 |
Hb_000212_440 |
0.0815019931 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_004096_240 |
0.0816998129 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |