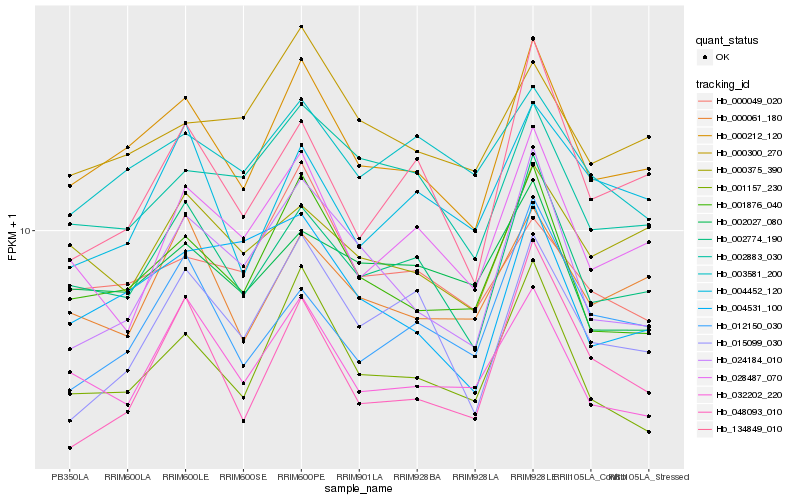
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 2 |
Hb_002774_190 |
0.0517214934 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 3 |
Hb_001876_040 |
0.0590767386 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 4 |
Hb_001157_230 |
0.0658091066 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 5 |
Hb_032202_220 |
0.0767083034 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_000061_180 |
0.077153681 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 7 |
Hb_002883_030 |
0.0818482812 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 8 |
Hb_000375_390 |
0.0826193927 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 9 |
Hb_000300_270 |
0.0828333166 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_024184_010 |
0.0840391685 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_028487_070 |
0.0860257516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000049_020 |
0.0865492382 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 13 |
Hb_002027_080 |
0.0869003074 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 14 |
Hb_134849_010 |
0.0870074055 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 15 |
Hb_004531_100 |
0.0876028653 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003581_200 |
0.087615767 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 17 |
Hb_015099_030 |
0.0884088457 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 18 |
Hb_012150_030 |
0.089787061 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_004452_120 |
0.0919839782 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 20 |
Hb_048093_010 |
0.0920346127 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |