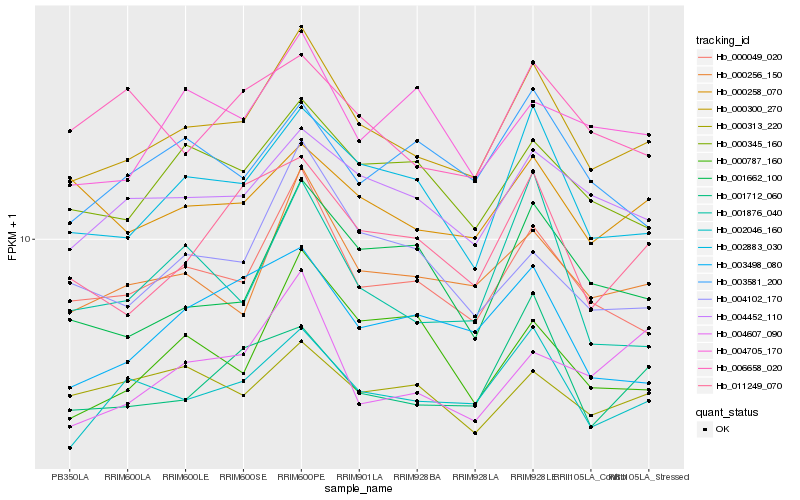
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_270 |
0.0 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_000049_020 |
0.0506830757 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 3 |
Hb_002046_160 |
0.0546163061 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000256_150 |
0.0558917111 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 5 |
Hb_002883_030 |
0.0559592359 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 6 |
Hb_004452_110 |
0.058030795 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000345_160 |
0.0642968555 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 8 |
Hb_011249_070 |
0.0673858544 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 9 |
Hb_001712_060 |
0.0686504649 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 10 |
Hb_003498_080 |
0.0737007857 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 11 |
Hb_001662_100 |
0.0743106054 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 12 |
Hb_000258_070 |
0.0746887503 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 13 |
Hb_004102_170 |
0.0764366856 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 14 |
Hb_004705_170 |
0.0781963694 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 15 |
Hb_004607_090 |
0.0793056874 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 16 |
Hb_000787_160 |
0.0797359939 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003581_200 |
0.0801801707 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 18 |
Hb_006658_020 |
0.0802109503 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 19 |
Hb_001876_040 |
0.0805480879 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 20 |
Hb_000313_220 |
0.0811899922 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |