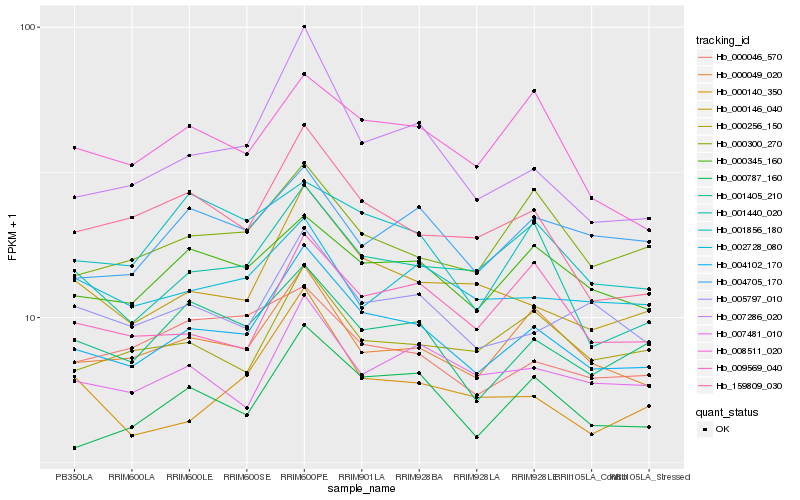
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004102_170 |
0.0 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 2 |
Hb_000787_160 |
0.0567447938 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000049_020 |
0.0611180699 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 4 |
Hb_000146_040 |
0.0635667146 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 5 |
Hb_000345_160 |
0.0639483386 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 6 |
Hb_001405_210 |
0.0646268997 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 7 |
Hb_005797_010 |
0.0693731173 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_007481_010 |
0.0750339817 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 9 |
Hb_159809_030 |
0.0757310126 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000256_150 |
0.0757392709 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 11 |
Hb_007286_020 |
0.0759863142 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 12 |
Hb_000300_270 |
0.0764366856 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000140_350 |
0.0782046834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001856_180 |
0.0784457367 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 15 |
Hb_001440_020 |
0.0785711437 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 16 |
Hb_000046_570 |
0.0802115046 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 17 |
Hb_009569_040 |
0.0806623702 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 18 |
Hb_004705_170 |
0.0806722102 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 19 |
Hb_002728_080 |
0.0811405277 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_008511_020 |
0.0819598178 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |