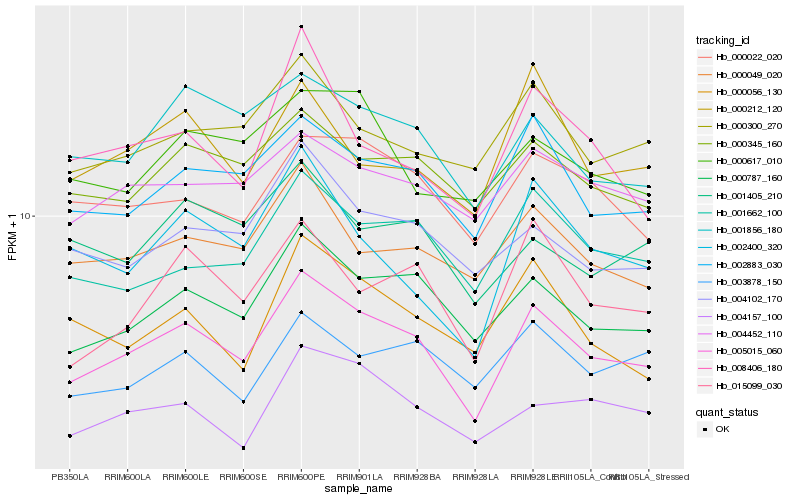
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005015_060 |
0.0 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Jatropha curcas] |
| 2 |
Hb_000787_160 |
0.0668859681 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002883_030 |
0.0732821203 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 4 |
Hb_015099_030 |
0.0841036925 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 5 |
Hb_001662_100 |
0.0857961484 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 6 |
Hb_001856_180 |
0.0863055331 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 7 |
Hb_000345_160 |
0.0865812805 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 8 |
Hb_000049_020 |
0.0880539503 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 9 |
Hb_004102_170 |
0.0884279155 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 10 |
Hb_000022_020 |
0.0884508688 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 11 |
Hb_000300_270 |
0.0898362032 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_004452_110 |
0.0905275203 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004157_100 |
0.0921798515 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 14 |
Hb_002400_320 |
0.0939450033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000212_120 |
0.0959074422 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 16 |
Hb_008406_180 |
0.0971373638 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_003878_150 |
0.097311696 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 18 |
Hb_000056_130 |
0.097465783 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 19 |
Hb_001405_210 |
0.0989171323 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 20 |
Hb_000617_010 |
0.0996446061 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |