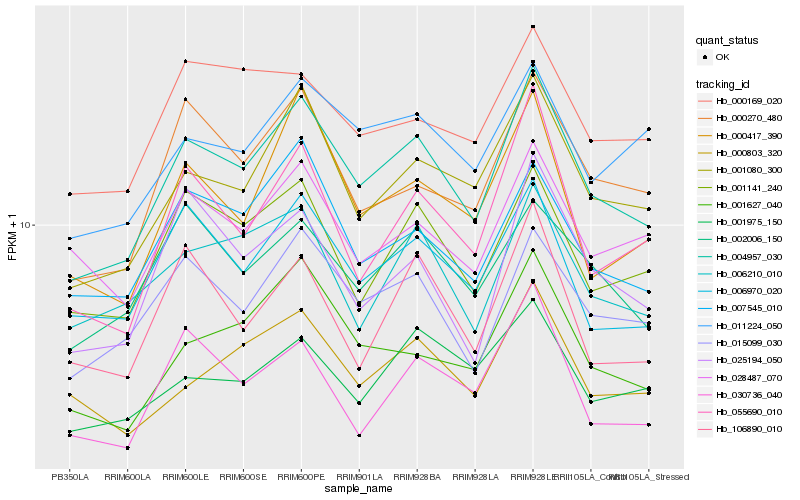
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004957_030 |
0.0 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 2 |
Hb_001080_300 |
0.0660708689 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 3 |
Hb_015099_030 |
0.0713707716 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 4 |
Hb_001141_240 |
0.0782737236 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_007545_010 |
0.0798686086 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 6 |
Hb_006970_020 |
0.0801652437 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 7 |
Hb_001627_040 |
0.0815775207 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000270_480 |
0.0821362309 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 9 |
Hb_002006_150 |
0.0852132785 |
- |
- |
copine, putative [Ricinus communis] |
| 10 |
Hb_006210_010 |
0.0863505355 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 11 |
Hb_055690_010 |
0.0869820453 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000417_390 |
0.0882489475 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 13 |
Hb_011224_050 |
0.0902627361 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Populus euphratica] |
| 14 |
Hb_000803_320 |
0.0906466795 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 15 |
Hb_025194_050 |
0.09170145 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000169_020 |
0.0938443278 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 17 |
Hb_028487_070 |
0.0949934752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001975_150 |
0.095531574 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 19 |
Hb_030736_040 |
0.0955896597 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 20 |
Hb_106890_010 |
0.0962051682 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |