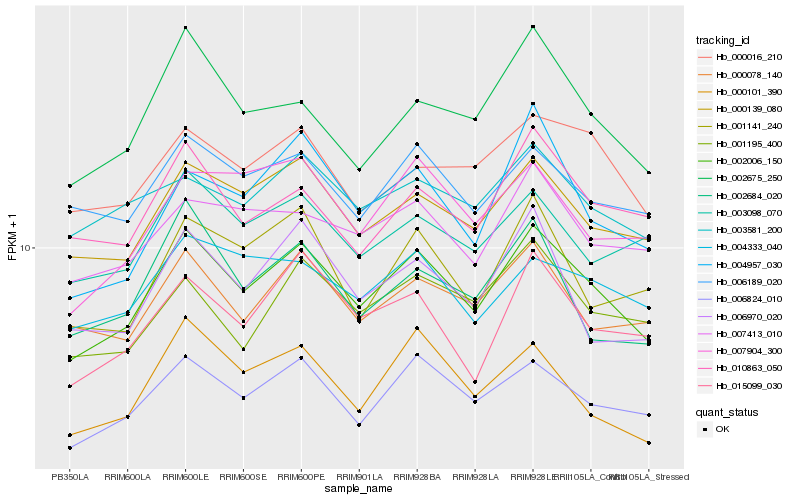
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002006_150 |
0.0 |
- |
- |
copine, putative [Ricinus communis] |
| 2 |
Hb_006824_010 |
0.0586914749 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 3 |
Hb_000139_080 |
0.0668278567 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000078_140 |
0.0791252874 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 5 |
Hb_015099_030 |
0.0791500656 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 6 |
Hb_002675_250 |
0.0803919366 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 7 |
Hb_000101_390 |
0.0823284288 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 8 |
Hb_006189_020 |
0.0824807251 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 9 |
Hb_007413_010 |
0.0827880662 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 10 |
Hb_006970_020 |
0.083534284 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 11 |
Hb_007904_300 |
0.0837712626 |
- |
- |
copine, putative [Ricinus communis] |
| 12 |
Hb_003098_070 |
0.0846783154 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 13 |
Hb_001195_400 |
0.0851156986 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 14 |
Hb_004957_030 |
0.0852132785 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 15 |
Hb_001141_240 |
0.0854397036 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004333_040 |
0.0858382316 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 17 |
Hb_000016_210 |
0.0862220125 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 18 |
Hb_010863_050 |
0.08648687 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 19 |
Hb_002684_020 |
0.0866760151 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 20 |
Hb_003581_200 |
0.0867254252 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |