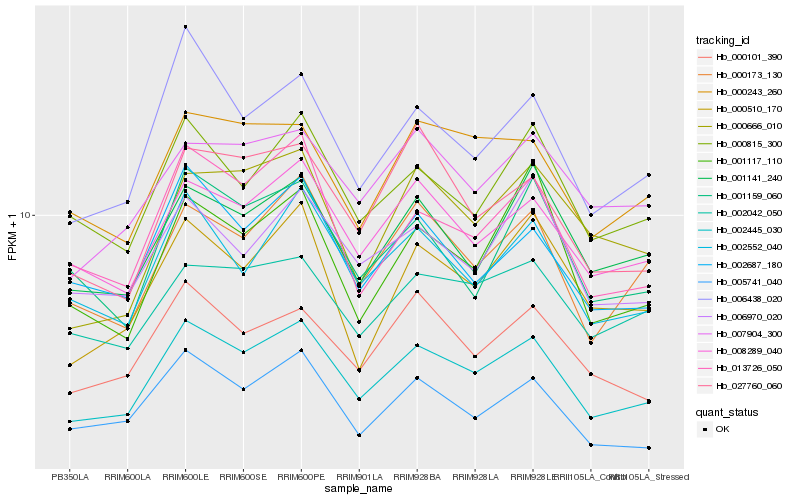
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002445_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 2 |
Hb_000173_130 |
0.0723441493 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006438_020 |
0.0727336309 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 4 |
Hb_013726_050 |
0.0734457679 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 5 |
Hb_008289_040 |
0.0747094404 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 6 |
Hb_002552_040 |
0.0763846772 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 7 |
Hb_002687_180 |
0.0812622743 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 8 |
Hb_000101_390 |
0.0817563744 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 9 |
Hb_000815_300 |
0.0834692581 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 10 |
Hb_001141_240 |
0.085087651 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000510_170 |
0.0859300167 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 12 |
Hb_005741_040 |
0.0864590006 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 13 |
Hb_000243_260 |
0.0878374289 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 14 |
Hb_027760_060 |
0.0881534235 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 15 |
Hb_000666_010 |
0.0896622484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006970_020 |
0.0897303796 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 17 |
Hb_007904_300 |
0.0905842448 |
- |
- |
copine, putative [Ricinus communis] |
| 18 |
Hb_001159_060 |
0.0907394206 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 19 |
Hb_001117_110 |
0.0914734117 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 20 |
Hb_002042_050 |
0.0916683112 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |