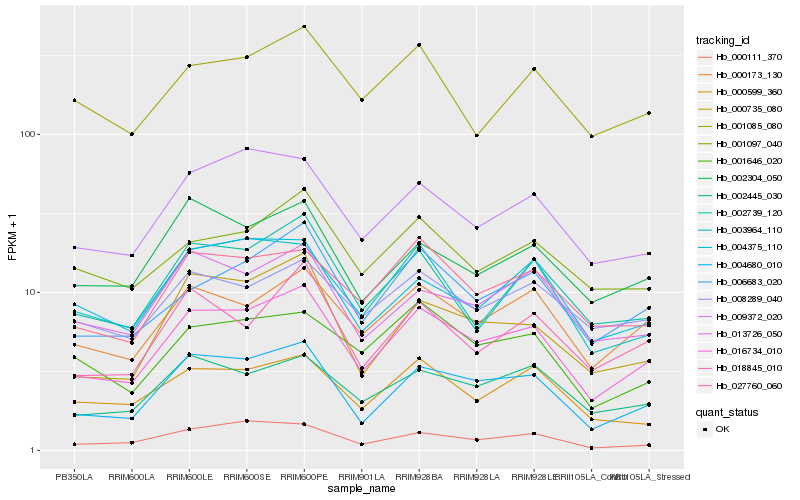
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016734_010 |
0.0 |
- |
- |
PREDICTED: probable DNA-3-methyladenine glycosylase 2 [Jatropha curcas] |
| 2 |
Hb_004680_010 |
0.0602316294 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006683_020 |
0.0863169295 |
- |
- |
PREDICTED: nucleosome assembly protein 1;4 [Jatropha curcas] |
| 4 |
Hb_000173_130 |
0.0863995962 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002445_030 |
0.0922608778 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 6 |
Hb_009372_020 |
0.0956204483 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 7 |
Hb_027760_060 |
0.0972843388 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 8 |
Hb_002739_120 |
0.0983377851 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 9 |
Hb_000735_080 |
0.0986003858 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 10 |
Hb_013726_050 |
0.1032204581 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 11 |
Hb_018845_010 |
0.1059577352 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 12 |
Hb_001097_040 |
0.1064194933 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 13 |
Hb_000599_360 |
0.1067498216 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 14 |
Hb_002304_050 |
0.1073108139 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 15 |
Hb_000111_370 |
0.1076872521 |
- |
- |
PREDICTED: cation/H(+) antiporter 2-like [Jatropha curcas] |
| 16 |
Hb_001646_020 |
0.1079405978 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004375_110 |
0.1081764904 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 18 |
Hb_008289_040 |
0.1085419481 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 19 |
Hb_003964_110 |
0.1086344963 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001085_080 |
0.1109705866 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |