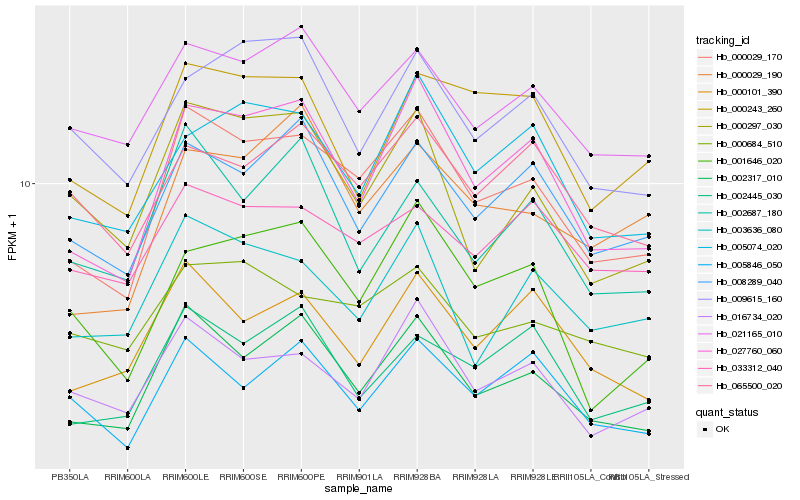
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000029_170 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 2 |
Hb_027760_060 |
0.0625065565 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 3 |
Hb_002317_010 |
0.0727622608 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_008289_040 |
0.0927317324 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 5 |
Hb_016734_020 |
0.0943476034 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 6 |
Hb_021165_010 |
0.0957640605 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_005074_020 |
0.0965664111 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000101_390 |
0.0995453668 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 9 |
Hb_000684_510 |
0.0998427208 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 10 |
Hb_000029_190 |
0.1025691456 |
- |
- |
hypothetical protein CISIN_1g025338mg [Citrus sinensis] |
| 11 |
Hb_001646_020 |
0.1033366772 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000297_030 |
0.1046171836 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002445_030 |
0.1048834841 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 14 |
Hb_003636_080 |
0.104889591 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 15 |
Hb_065500_020 |
0.1050343591 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 16 |
Hb_005846_050 |
0.1058875721 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 17 |
Hb_033312_040 |
0.1089009287 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000243_260 |
0.1093500754 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009615_160 |
0.109869453 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 20 |
Hb_002687_180 |
0.1102085539 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |