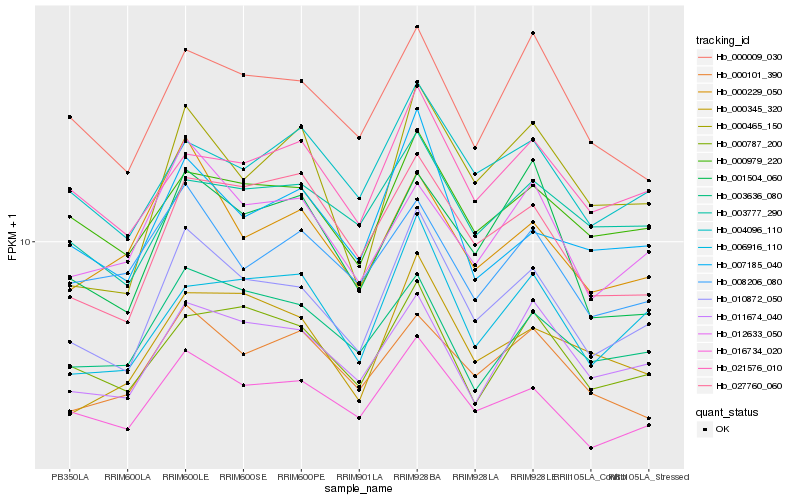
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010872_050 |
0.0 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 2 |
Hb_016734_020 |
0.0695738189 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_007185_040 |
0.0984194624 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 4 |
Hb_011674_040 |
0.0989873922 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006916_110 |
0.099247955 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000979_220 |
0.100498685 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000345_320 |
0.1023623199 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_027760_060 |
0.1041147188 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 9 |
Hb_000787_200 |
0.1043411318 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000465_150 |
0.1054107307 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 11 |
Hb_004096_110 |
0.1060702775 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 12 |
Hb_003636_080 |
0.1067307959 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 13 |
Hb_021576_010 |
0.1073513421 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 14 |
Hb_012633_050 |
0.1077658925 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_000229_050 |
0.108389789 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 16 |
Hb_000101_390 |
0.1084277005 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 17 |
Hb_000009_030 |
0.1084935515 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 18 |
Hb_003777_290 |
0.1093167849 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 19 |
Hb_001504_060 |
0.1116423144 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 20 |
Hb_008206_080 |
0.1153530969 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 17 [Jatropha curcas] |