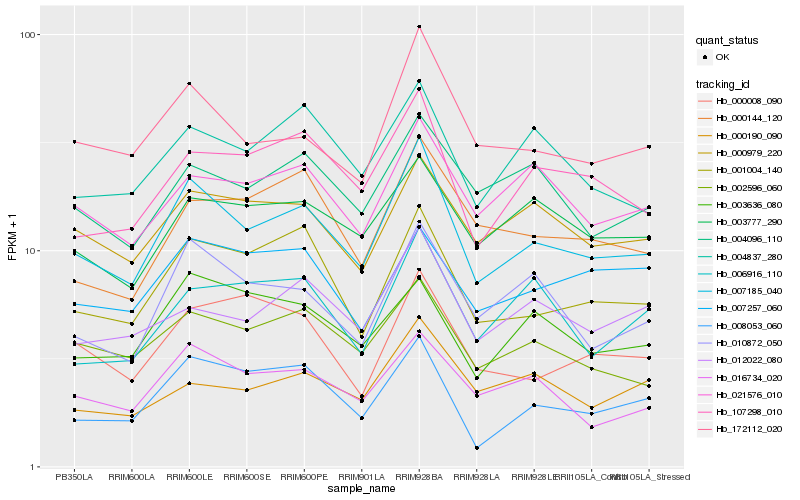
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007185_040 |
0.0 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 2 |
Hb_172112_020 |
0.0849649509 |
- |
- |
PREDICTED: fumarylacetoacetase [Jatropha curcas] |
| 3 |
Hb_001004_140 |
0.0920648237 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 4 |
Hb_002596_060 |
0.0952007759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010872_050 |
0.0984194624 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 6 |
Hb_016734_020 |
0.0984512339 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_107298_010 |
0.0995452831 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 8 |
Hb_000979_220 |
0.1020422476 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 9 |
Hb_021576_010 |
0.105759243 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 10 |
Hb_004837_280 |
0.107622769 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 11 |
Hb_008053_060 |
0.1079430203 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_012022_080 |
0.108998513 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_003777_290 |
0.109666175 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 14 |
Hb_004096_110 |
0.1108419133 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 15 |
Hb_000144_120 |
0.1137337817 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_003636_080 |
0.116816752 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 17 |
Hb_000190_090 |
0.1197928291 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 18 |
Hb_000008_090 |
0.1199246377 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 19 |
Hb_007257_060 |
0.1220384538 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 20 |
Hb_006916_110 |
0.1220766592 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |