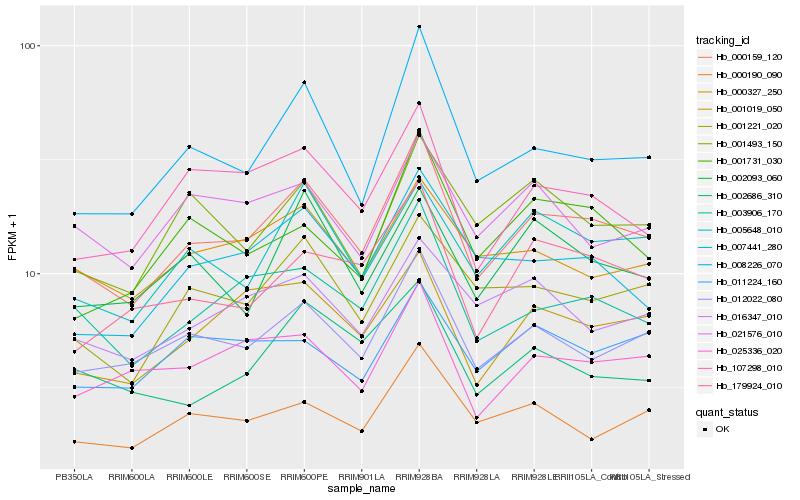
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000159_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 2 |
Hb_012022_080 |
0.0759697711 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_008226_070 |
0.0809999299 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 4 |
Hb_003906_170 |
0.0823793176 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_001019_050 |
0.0918017797 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 6 |
Hb_001221_020 |
0.0926080938 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 7 |
Hb_107298_010 |
0.0928541357 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 8 |
Hb_005648_010 |
0.0941457816 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 9 |
Hb_007441_280 |
0.0971597039 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 10 |
Hb_000190_090 |
0.0978873683 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 11 |
Hb_179924_010 |
0.0986398873 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_025336_020 |
0.1016018184 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 13 |
Hb_000327_250 |
0.1020500131 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 14 |
Hb_002093_060 |
0.1046708307 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 15 |
Hb_001493_150 |
0.1052527793 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 16 |
Hb_016347_010 |
0.1054866476 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 17 |
Hb_001731_030 |
0.1062887922 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A-like [Gossypium raimondii] |
| 18 |
Hb_002686_310 |
0.1086093813 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 19 |
Hb_021576_010 |
0.1089075582 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 20 |
Hb_011224_160 |
0.1100338203 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |