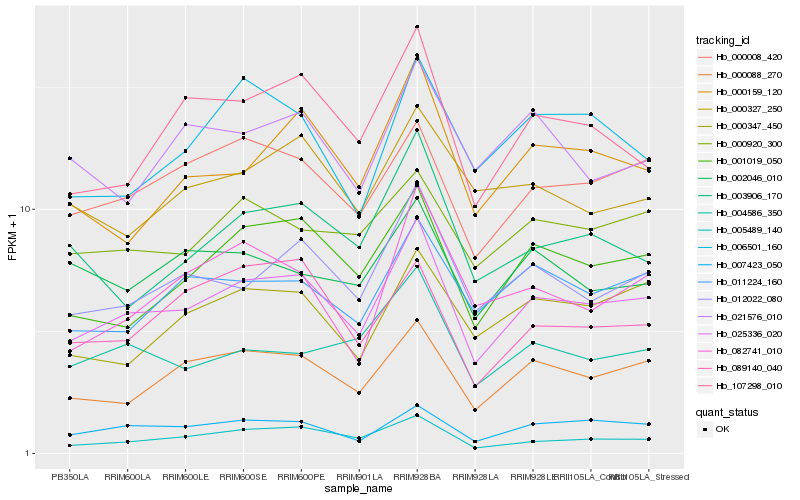
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025336_020 |
0.0 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 2 |
Hb_000088_270 |
0.0779537102 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 3 |
Hb_007423_050 |
0.0787473896 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 3 [Jatropha curcas] |
| 4 |
Hb_012022_080 |
0.0818342834 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_001019_050 |
0.0847554017 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 6 |
Hb_000008_420 |
0.0861151292 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005489_140 |
0.0875025096 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 8 |
Hb_107298_010 |
0.0957331829 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 9 |
Hb_011224_160 |
0.0961762692 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 10 |
Hb_000159_120 |
0.1016018184 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 11 |
Hb_000347_450 |
0.103636512 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000920_300 |
0.104073812 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 13 |
Hb_004586_350 |
0.1078059268 |
- |
- |
Phosphoribosylaminoimidazole-succinocarboxamide synthase [Morus notabilis] |
| 14 |
Hb_003906_170 |
0.1089280231 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_006501_160 |
0.1094964775 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 16 |
Hb_082741_010 |
0.1130103531 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 17 |
Hb_000327_250 |
0.1139697462 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 18 |
Hb_021576_010 |
0.1140966884 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 19 |
Hb_002046_010 |
0.1144006599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_089140_040 |
0.1159145534 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |