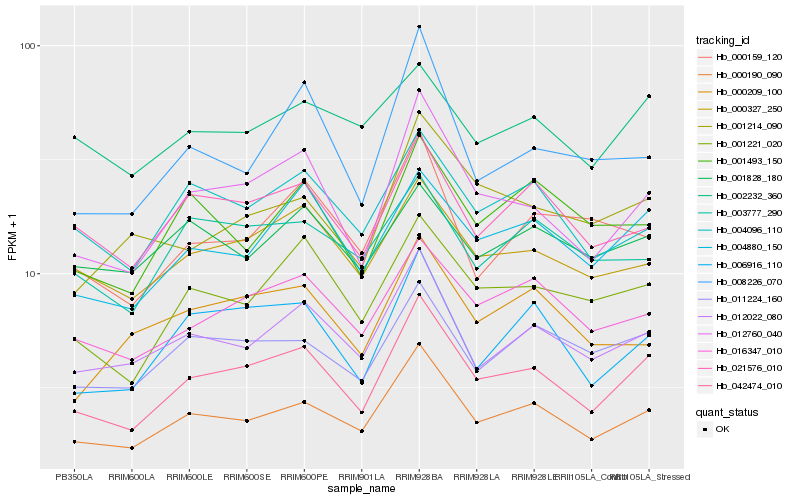
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000190_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 2 |
Hb_012022_080 |
0.0561232652 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_042474_010 |
0.0593293942 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 4 |
Hb_004096_110 |
0.0782734878 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 5 |
Hb_011224_160 |
0.0796213259 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 6 |
Hb_021576_010 |
0.0802982846 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 7 |
Hb_016347_010 |
0.0809815017 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 8 |
Hb_012760_040 |
0.0864578711 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 9 |
Hb_000327_250 |
0.0892197972 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 10 |
Hb_001493_150 |
0.0900906836 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 11 |
Hb_003777_290 |
0.0932657211 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 12 |
Hb_006916_110 |
0.0934878536 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004880_150 |
0.0939578706 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 14 |
Hb_001221_020 |
0.0942967477 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 15 |
Hb_000159_120 |
0.0978873683 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 16 |
Hb_002232_360 |
0.1006701119 |
- |
- |
PREDICTED: proline iminopeptidase isoform X2 [Jatropha curcas] |
| 17 |
Hb_001828_180 |
0.1007215137 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 18 |
Hb_000209_100 |
0.1042852066 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001214_090 |
0.1055829945 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 20 |
Hb_008226_070 |
0.1057160168 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |