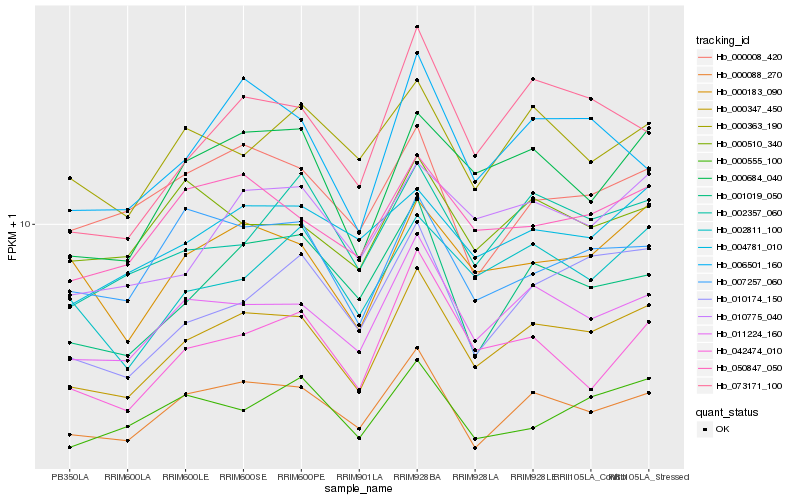
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_450 |
0.0 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_011224_160 |
0.0621194274 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 3 |
Hb_000088_270 |
0.0690977585 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 4 |
Hb_000684_040 |
0.075943096 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 5 |
Hb_050847_050 |
0.084741579 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010775_040 |
0.086525147 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 7 |
Hb_000183_090 |
0.0873509088 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X2 [Jatropha curcas] |
| 8 |
Hb_010174_150 |
0.090559534 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_001019_050 |
0.0915844993 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 10 |
Hb_002357_060 |
0.0917878103 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 11 |
Hb_073171_100 |
0.0918699701 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 12 |
Hb_002811_100 |
0.0934969257 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 13 |
Hb_006501_160 |
0.0957688074 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 14 |
Hb_004781_010 |
0.0967384619 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 15 |
Hb_042474_010 |
0.1001198133 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 16 |
Hb_007257_060 |
0.1006284078 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 17 |
Hb_000008_420 |
0.1009916699 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000363_190 |
0.1021152095 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000555_100 |
0.1031070859 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 20 |
Hb_000510_340 |
0.1036169661 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |