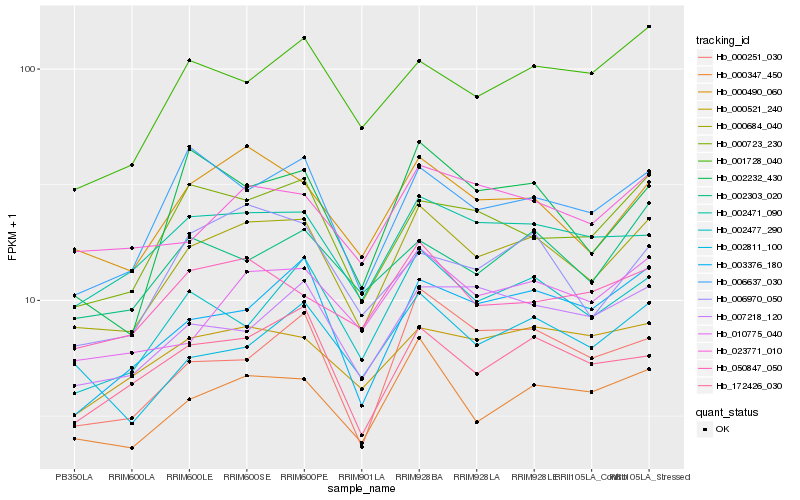
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_040 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 2 |
Hb_000490_060 |
0.07385314 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 3 |
Hb_000347_450 |
0.075943096 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_002471_090 |
0.0862798216 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 5 |
Hb_000723_230 |
0.0868389126 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 6 |
Hb_010775_040 |
0.0884914112 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 7 |
Hb_002477_290 |
0.0899855733 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_006637_030 |
0.0908209073 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 9 |
Hb_003376_180 |
0.0917859466 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 10 |
Hb_172426_030 |
0.0919829472 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 11 |
Hb_023771_010 |
0.0949136106 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 12 |
Hb_002811_100 |
0.0959918754 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 13 |
Hb_006970_050 |
0.0973592122 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 14 |
Hb_000521_240 |
0.097882779 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001728_040 |
0.0980104367 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 16 |
Hb_002303_020 |
0.0980985136 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Gossypium raimondii] |
| 17 |
Hb_007218_120 |
0.0981337213 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002232_430 |
0.0986736234 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 19 |
Hb_050847_050 |
0.0990757506 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000251_030 |
0.1002979797 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |