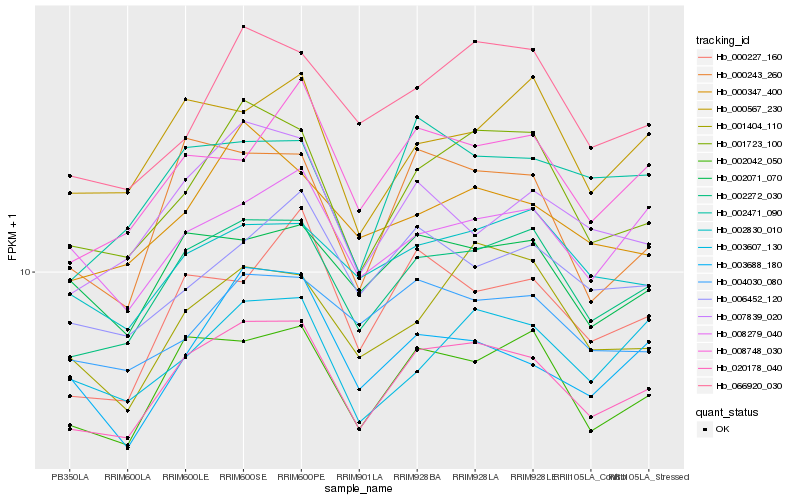
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020178_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002272_030 |
0.0375054593 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 3 |
Hb_002042_050 |
0.0525924528 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001723_100 |
0.0605115399 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002071_070 |
0.0700329333 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000243_260 |
0.0727184439 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000347_400 |
0.0732243001 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008748_030 |
0.0740939404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002830_010 |
0.0749442257 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 10 |
Hb_004030_080 |
0.0764823921 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 11 |
Hb_066920_030 |
0.0772610527 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 12 |
Hb_007839_020 |
0.077281399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002471_090 |
0.0785083414 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 14 |
Hb_006452_120 |
0.0785126499 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 15 |
Hb_008279_040 |
0.079078557 |
- |
- |
Rer1 family protein isoform 2 [Theobroma cacao] |
| 16 |
Hb_000227_160 |
0.0793167094 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 17 |
Hb_000567_230 |
0.0795899539 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 18 |
Hb_001404_110 |
0.0806747185 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 19 |
Hb_003688_180 |
0.0809479009 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 20 |
Hb_003607_130 |
0.0811683649 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |