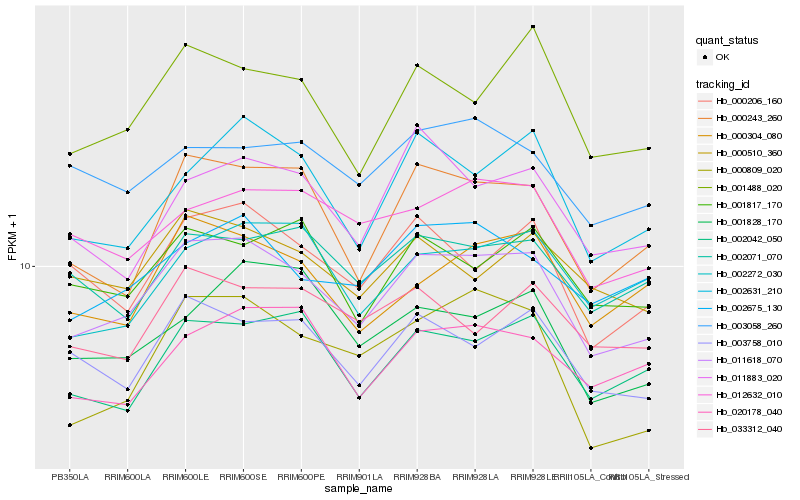
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011618_070 |
0.0 |
- |
- |
NMDA receptor-regulated protein [Medicago truncatula] |
| 2 |
Hb_000243_260 |
0.0714654308 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001488_020 |
0.0751643368 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 4 |
Hb_003758_010 |
0.0760488744 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002631_210 |
0.0766630158 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 6 |
Hb_002042_050 |
0.0767967809 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000206_160 |
0.0769590614 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_000304_080 |
0.0800752348 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002675_130 |
0.0802766698 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 16, NatA auxiliary subunit isoform X1 [Jatropha curcas] |
| 10 |
Hb_012632_010 |
0.0838395427 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 11 |
Hb_000510_360 |
0.0848652562 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 12 |
Hb_002272_030 |
0.0850488593 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 13 |
Hb_001817_170 |
0.0856470815 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 14 |
Hb_011883_020 |
0.0862371345 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 15 |
Hb_020178_040 |
0.0863575839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002071_070 |
0.0864638639 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003058_260 |
0.0896881289 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 18 |
Hb_033312_040 |
0.0897349515 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000809_020 |
0.0899182041 |
desease resistance |
Gene Name: RPW8 |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001828_170 |
0.0907573896 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |