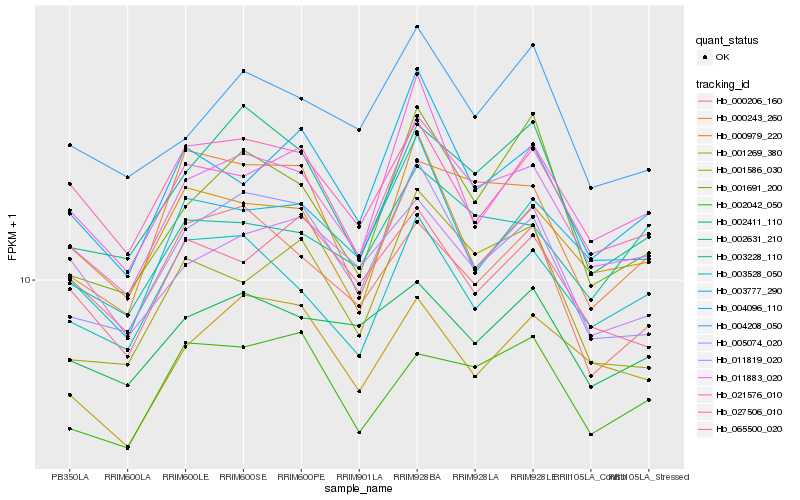
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011883_020 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 2 |
Hb_005074_020 |
0.0534425208 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002631_210 |
0.0631402371 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 4 |
Hb_027506_010 |
0.0712379969 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 5 |
Hb_004208_050 |
0.0719960042 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 6 |
Hb_004096_110 |
0.0734671952 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 7 |
Hb_001691_200 |
0.0740457037 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 8 |
Hb_001586_030 |
0.0743658111 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 9 |
Hb_003777_290 |
0.0745273411 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 10 |
Hb_000979_220 |
0.0747294412 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000243_260 |
0.0748027064 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003528_050 |
0.0752730051 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 13 |
Hb_001269_380 |
0.0767001866 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 14 |
Hb_021576_010 |
0.0785595904 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 15 |
Hb_002042_050 |
0.0792602312 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 16 |
Hb_011819_020 |
0.079511135 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002411_110 |
0.0795727105 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 18 |
Hb_003228_110 |
0.0801247257 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 19 |
Hb_065500_020 |
0.0811052648 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 20 |
Hb_000206_160 |
0.0811532287 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |