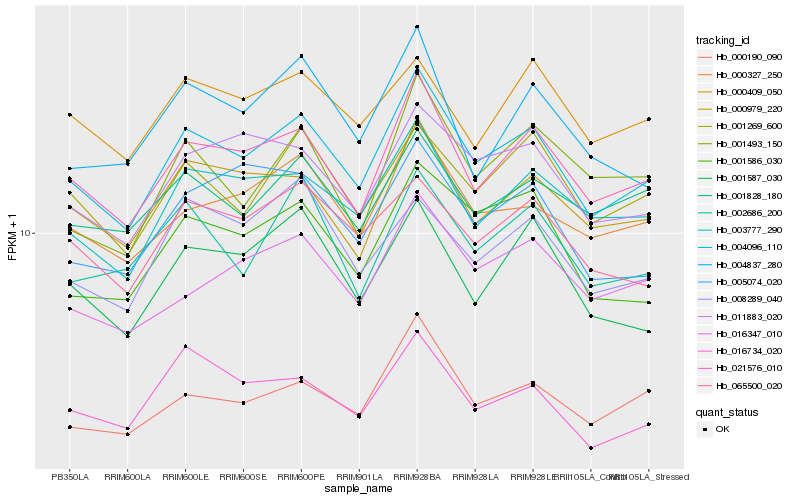
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004096_110 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 2 |
Hb_021576_010 |
0.0452133677 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 3 |
Hb_003777_290 |
0.0629049436 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 4 |
Hb_000327_250 |
0.0660376411 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 5 |
Hb_000979_220 |
0.0707805928 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011883_020 |
0.0734671952 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 7 |
Hb_001269_600 |
0.073905074 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 8 |
Hb_065500_020 |
0.075240168 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 9 |
Hb_016347_010 |
0.0773798968 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 10 |
Hb_001586_030 |
0.0776512612 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 11 |
Hb_000190_090 |
0.0782734878 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 12 |
Hb_001828_180 |
0.0782804627 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 13 |
Hb_001587_030 |
0.081793606 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 14 |
Hb_002686_200 |
0.084857422 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 15 |
Hb_005074_020 |
0.0858740648 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004837_280 |
0.0863137107 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 17 |
Hb_000409_050 |
0.0870972145 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 18 |
Hb_008289_040 |
0.0871892959 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 19 |
Hb_001493_150 |
0.0873469658 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 20 |
Hb_016734_020 |
0.0874335165 |
- |
- |
sugar transporter, putative [Ricinus communis] |