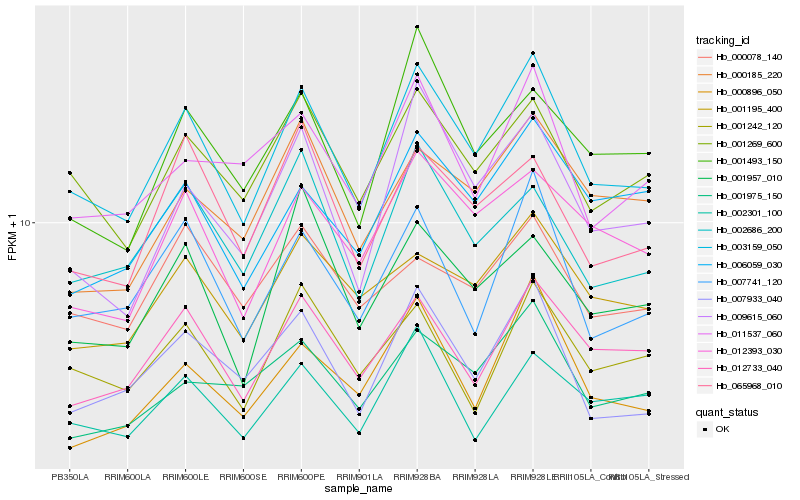
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003159_050 |
0.0 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_001269_600 |
0.0680474648 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_007933_040 |
0.0715181916 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 4 |
Hb_012733_040 |
0.0735073807 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 5 |
Hb_007741_120 |
0.0745658365 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_012393_030 |
0.0745767895 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 7 |
Hb_009615_060 |
0.0784788821 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 8 |
Hb_001195_400 |
0.0792996217 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 9 |
Hb_002686_200 |
0.079782602 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 10 |
Hb_065968_010 |
0.0845672965 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002301_100 |
0.0847374114 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 12 |
Hb_000896_050 |
0.0861617058 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |
| 13 |
Hb_001493_150 |
0.0872628106 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 14 |
Hb_011537_060 |
0.0873656166 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 15 |
Hb_001975_150 |
0.0875179287 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 16 |
Hb_006059_030 |
0.0882381633 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_001242_120 |
0.088512935 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 18 |
Hb_001957_010 |
0.0885883298 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000078_140 |
0.0899323742 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 20 |
Hb_000185_220 |
0.0908382597 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |