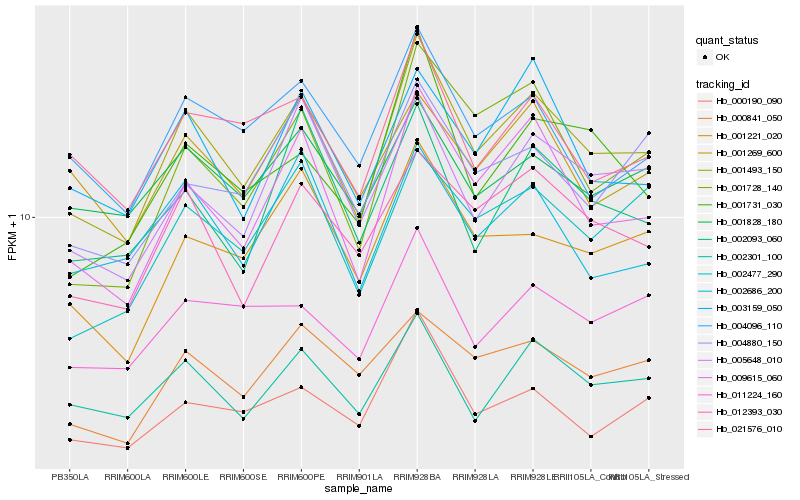
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001493_150 |
0.0 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 2 |
Hb_009615_060 |
0.0617381466 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 3 |
Hb_002301_100 |
0.0766277477 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 4 |
Hb_012393_030 |
0.077906765 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 5 |
Hb_001221_020 |
0.0837646528 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 6 |
Hb_005648_010 |
0.0840828283 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 7 |
Hb_021576_010 |
0.0868239996 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 8 |
Hb_002477_290 |
0.0868637635 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_003159_050 |
0.0872628106 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_004096_110 |
0.0873469658 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 11 |
Hb_001728_140 |
0.0875745833 |
- |
- |
- |
| 12 |
Hb_000841_050 |
0.0892145838 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 13 |
Hb_000190_090 |
0.0900906836 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 14 |
Hb_001828_180 |
0.0902492154 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 15 |
Hb_011224_160 |
0.090326995 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 16 |
Hb_002686_200 |
0.0923261228 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 17 |
Hb_001731_030 |
0.0932402874 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A-like [Gossypium raimondii] |
| 18 |
Hb_002093_060 |
0.0937842646 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 19 |
Hb_001269_600 |
0.0945180315 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 20 |
Hb_004880_150 |
0.0961176663 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |