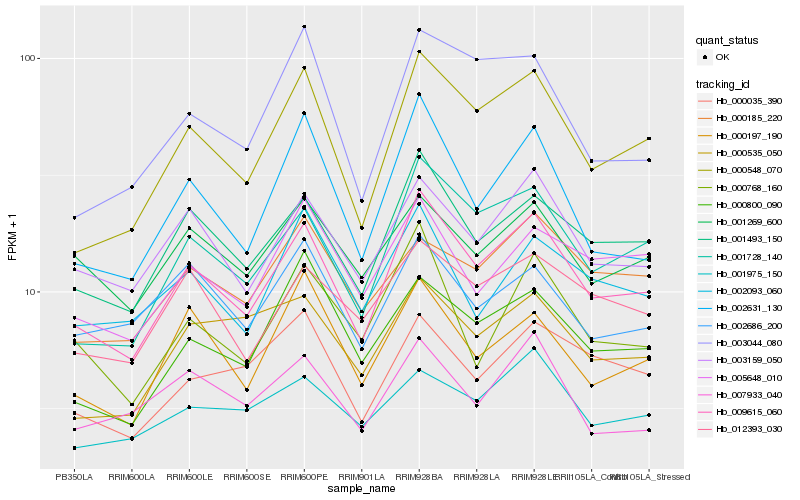
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009615_060 |
0.0 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 2 |
Hb_001493_150 |
0.0617381466 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 3 |
Hb_000548_070 |
0.0654817236 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 4 |
Hb_001728_140 |
0.0675193433 |
- |
- |
- |
| 5 |
Hb_003159_050 |
0.0784788821 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_002631_130 |
0.0882648465 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 7 |
Hb_012393_030 |
0.0956673824 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 8 |
Hb_000197_190 |
0.0967966446 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000800_090 |
0.0992532958 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 10 |
Hb_005648_010 |
0.099770336 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 11 |
Hb_000768_160 |
0.1001227902 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 12 |
Hb_000035_390 |
0.1008127173 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 13 |
Hb_001269_600 |
0.1017704442 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_002093_060 |
0.1025808917 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 15 |
Hb_000185_220 |
0.1034708534 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 16 |
Hb_002686_200 |
0.1042442124 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 17 |
Hb_003044_080 |
0.104850611 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 18 |
Hb_007933_040 |
0.105017611 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000535_050 |
0.1052284222 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 20 |
Hb_001975_150 |
0.1065783657 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |