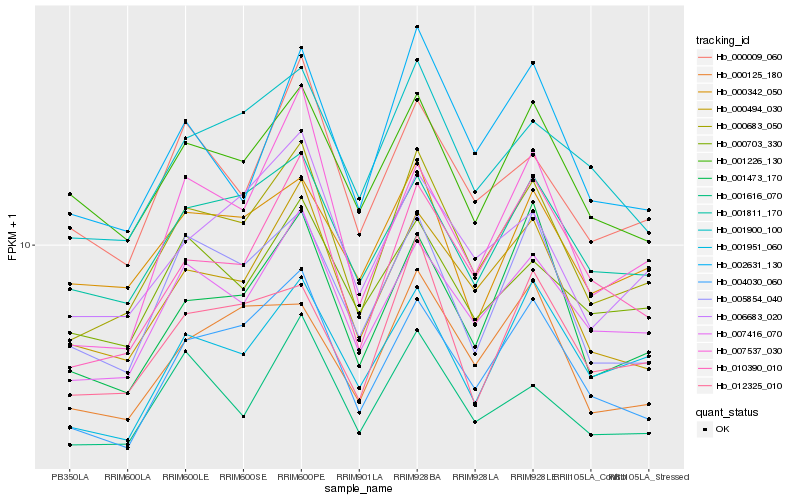
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000683_050 |
0.0 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 2 |
Hb_007416_070 |
0.0670818488 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 3 |
Hb_000494_030 |
0.0798980745 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 4 |
Hb_004030_060 |
0.0811474776 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 5 |
Hb_001811_170 |
0.0889611355 |
- |
- |
dynamin, putative [Ricinus communis] |
| 6 |
Hb_010390_010 |
0.0897313137 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 7 |
Hb_005854_040 |
0.0914540573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001900_100 |
0.094893753 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006683_020 |
0.0951532839 |
- |
- |
PREDICTED: nucleosome assembly protein 1;4 [Jatropha curcas] |
| 10 |
Hb_001616_070 |
0.0978064485 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 11 |
Hb_001951_060 |
0.0981341709 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 12 |
Hb_001226_130 |
0.0984635334 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 13 |
Hb_000009_060 |
0.1020720387 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 14 |
Hb_001473_170 |
0.102468401 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 15 |
Hb_012325_010 |
0.1031052975 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 16 |
Hb_007537_030 |
0.1038666082 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000703_330 |
0.1042635893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000342_050 |
0.1047164386 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 19 |
Hb_002631_130 |
0.1051984015 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 20 |
Hb_000125_180 |
0.1066792705 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |