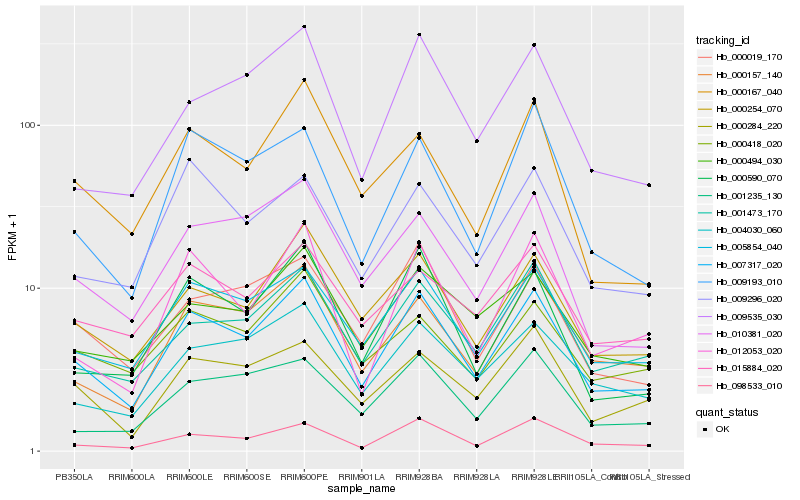
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007317_020 |
0.0 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005854_040 |
0.0798957201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000254_070 |
0.0872623712 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 4 |
Hb_000157_140 |
0.0927827097 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_098533_010 |
0.1005680381 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 6 |
Hb_010381_020 |
0.1009331611 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 7 |
Hb_001473_170 |
0.1010826101 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 8 |
Hb_000590_070 |
0.1041046086 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 9 |
Hb_000019_170 |
0.104312641 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 10 |
Hb_004030_060 |
0.1063091104 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 11 |
Hb_012053_020 |
0.1070947321 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000284_220 |
0.1073191884 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 13 |
Hb_000494_030 |
0.1094415939 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 14 |
Hb_009193_010 |
0.1112918532 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 15 |
Hb_000167_040 |
0.1129470341 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 16 |
Hb_009535_030 |
0.1138233826 |
- |
- |
CP2 [Hevea brasiliensis] |
| 17 |
Hb_015884_020 |
0.1154413203 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 18 |
Hb_000418_020 |
0.1155887929 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001235_130 |
0.1160294653 |
- |
- |
- |
| 20 |
Hb_009296_020 |
0.1165695109 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |