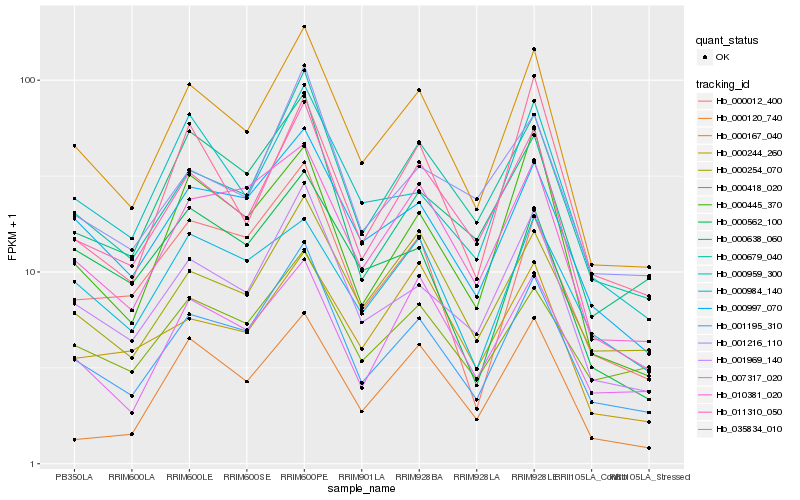
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000167_040 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 2 |
Hb_011310_050 |
0.0664393098 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 3 |
Hb_001195_310 |
0.0679852091 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_001969_140 |
0.0769452486 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 5 |
Hb_000997_070 |
0.0873486142 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 6 |
Hb_000679_040 |
0.0887600274 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 7 |
Hb_010381_020 |
0.0960812522 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 8 |
Hb_000959_300 |
0.1010940381 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 9 |
Hb_000254_070 |
0.1062285984 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 10 |
Hb_000445_370 |
0.1084318242 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 11 |
Hb_007317_020 |
0.1129470341 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000244_260 |
0.120749168 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000638_060 |
0.1208252916 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 14 |
Hb_000418_020 |
0.1210506139 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001216_110 |
0.1214307678 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 16 |
Hb_035834_010 |
0.1221417849 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_000984_140 |
0.123257878 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 18 |
Hb_000012_400 |
0.1236879668 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 19 |
Hb_000562_100 |
0.1238316433 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 20 |
Hb_000120_740 |
0.1240434461 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |