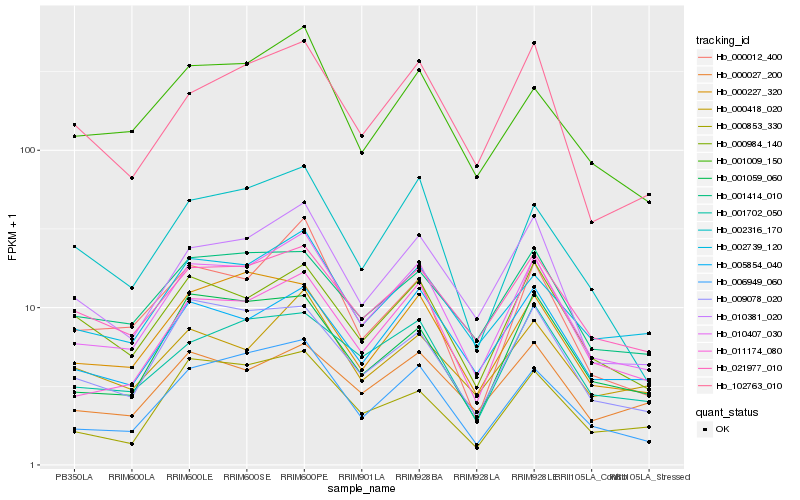
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010407_030 |
0.0 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 2 |
Hb_006949_060 |
0.06854752 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 3 |
Hb_010381_020 |
0.0818877727 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 4 |
Hb_002739_120 |
0.0851986203 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 5 |
Hb_001009_150 |
0.0911678378 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 6 |
Hb_005854_040 |
0.0919402632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009078_020 |
0.092369563 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 8 |
Hb_021977_010 |
0.0930751138 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 9 |
Hb_001059_060 |
0.1000322221 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 10 |
Hb_001702_050 |
0.1001887303 |
- |
- |
PREDICTED: probable protein S-acyltransferase 19 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000227_320 |
0.1002530286 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000984_140 |
0.1006544521 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 13 |
Hb_000012_400 |
0.1019245014 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 14 |
Hb_000853_330 |
0.1028086481 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 15 |
Hb_001414_010 |
0.1037366847 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 16 |
Hb_102763_010 |
0.104984576 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 17 |
Hb_002316_170 |
0.1067203595 |
- |
- |
purine permease, putative [Ricinus communis] |
| 18 |
Hb_000418_020 |
0.1067684743 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000027_200 |
0.1076255784 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 20 |
Hb_011174_080 |
0.1093692001 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |