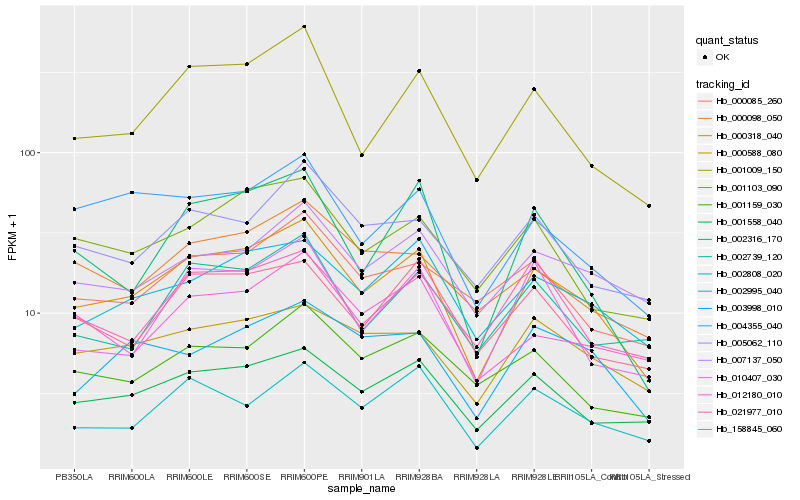
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_04877 [Jatropha curcas] |
| 2 |
Hb_001558_040 |
0.0940931608 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 3 |
Hb_001009_150 |
0.0946913323 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 4 |
Hb_002995_040 |
0.1038793869 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001103_090 |
0.1125127887 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 6 |
Hb_004355_040 |
0.1134749352 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 7 |
Hb_007137_050 |
0.1149164698 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_002808_020 |
0.1150456201 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X1 [Jatropha curcas] |
| 9 |
Hb_158845_060 |
0.1161889363 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002316_170 |
0.1164955631 |
- |
- |
purine permease, putative [Ricinus communis] |
| 11 |
Hb_000588_080 |
0.1167351395 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 12 |
Hb_005062_110 |
0.1174327257 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 13 |
Hb_012180_010 |
0.1194419271 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |
| 14 |
Hb_001159_030 |
0.1204200995 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 15 |
Hb_003998_010 |
0.1204557378 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 16 |
Hb_000098_050 |
0.1208939275 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 17 |
Hb_002739_120 |
0.1210491991 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 18 |
Hb_010407_030 |
0.1213642244 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 19 |
Hb_000085_260 |
0.1220412042 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 20 |
Hb_021977_010 |
0.123027431 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |