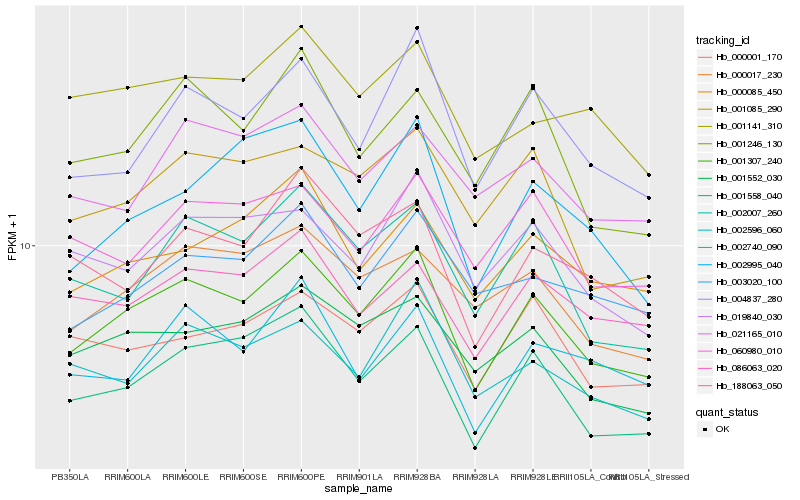
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001307_240 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001558_040 |
0.0666361191 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 3 |
Hb_004837_280 |
0.0754000001 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 4 |
Hb_002007_260 |
0.0776637014 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 5 |
Hb_001552_030 |
0.0795384223 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001085_290 |
0.0832834851 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 7 |
Hb_000017_230 |
0.0832881402 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 8 |
Hb_001246_130 |
0.0863688969 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000001_170 |
0.0868725434 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_019840_030 |
0.0873874985 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002596_060 |
0.0881088409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_188063_050 |
0.0881991495 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 13 |
Hb_000085_450 |
0.0884730146 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 14 |
Hb_002995_040 |
0.0900650425 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_086063_020 |
0.0908222084 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 16 |
Hb_060980_010 |
0.0917876074 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_002740_090 |
0.0930497946 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 18 |
Hb_003020_100 |
0.093777597 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 19 |
Hb_001141_310 |
0.0948033899 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 20 |
Hb_021165_010 |
0.095489232 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |