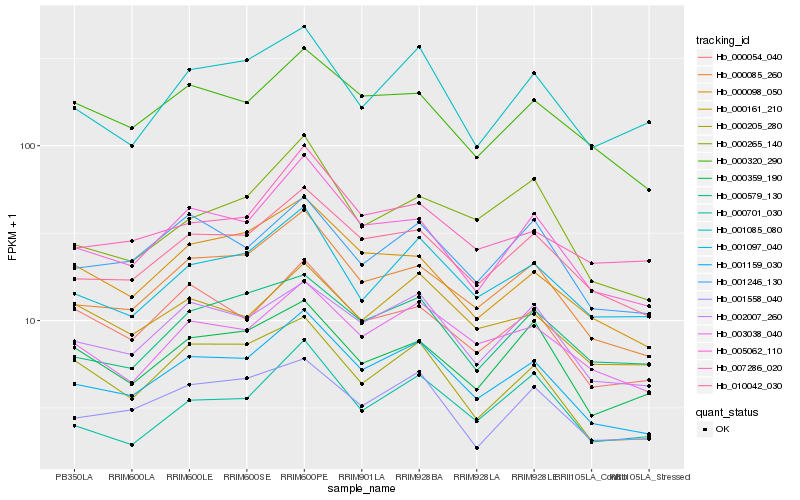
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001159_030 |
0.0 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 2 |
Hb_000085_260 |
0.0512081761 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 3 |
Hb_010042_030 |
0.0582811568 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 4 |
Hb_005062_110 |
0.0635041286 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 5 |
Hb_002007_260 |
0.0658225757 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 6 |
Hb_003038_040 |
0.0733575886 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 7 |
Hb_001097_040 |
0.0783661316 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 8 |
Hb_000320_290 |
0.0810432718 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 9 |
Hb_000098_050 |
0.0837435716 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 10 |
Hb_000054_040 |
0.0840620744 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 11 |
Hb_001558_040 |
0.0844765531 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 12 |
Hb_001246_130 |
0.0850261414 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000205_280 |
0.0866749462 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_000161_210 |
0.0884294327 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 15 |
Hb_001085_080 |
0.0904214622 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 16 |
Hb_000579_130 |
0.0922310313 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_000265_140 |
0.0929625 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 18 |
Hb_000701_030 |
0.0933073817 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 19 |
Hb_007286_020 |
0.0944269919 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 20 |
Hb_000359_190 |
0.094504732 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Nicotiana sylvestris] |