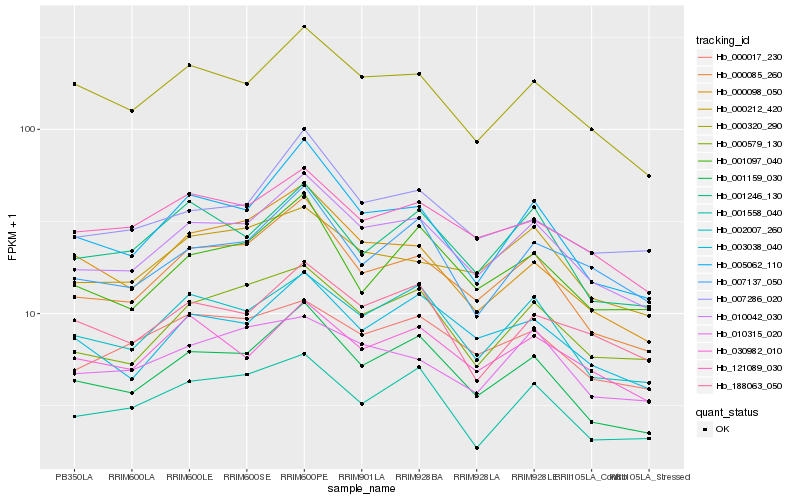
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010042_030 |
0.0 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 2 |
Hb_000085_260 |
0.0513127689 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 3 |
Hb_001159_030 |
0.0582811568 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 4 |
Hb_005062_110 |
0.062759746 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 5 |
Hb_002007_260 |
0.065609136 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 6 |
Hb_000579_130 |
0.0667235144 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_000320_290 |
0.068060121 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 8 |
Hb_003038_040 |
0.0737536137 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 9 |
Hb_121089_030 |
0.0759439355 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 10 |
Hb_000212_420 |
0.0764266559 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 11 |
Hb_007137_050 |
0.0778021487 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_007286_020 |
0.079590372 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 13 |
Hb_000017_230 |
0.0800166227 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 14 |
Hb_030982_010 |
0.0806235366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001246_130 |
0.0840885031 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 16 |
Hb_188063_050 |
0.0842516629 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 17 |
Hb_001097_040 |
0.085095079 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 18 |
Hb_000098_050 |
0.0851314126 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 19 |
Hb_010315_020 |
0.0852833151 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 20 |
Hb_001558_040 |
0.0865260119 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |