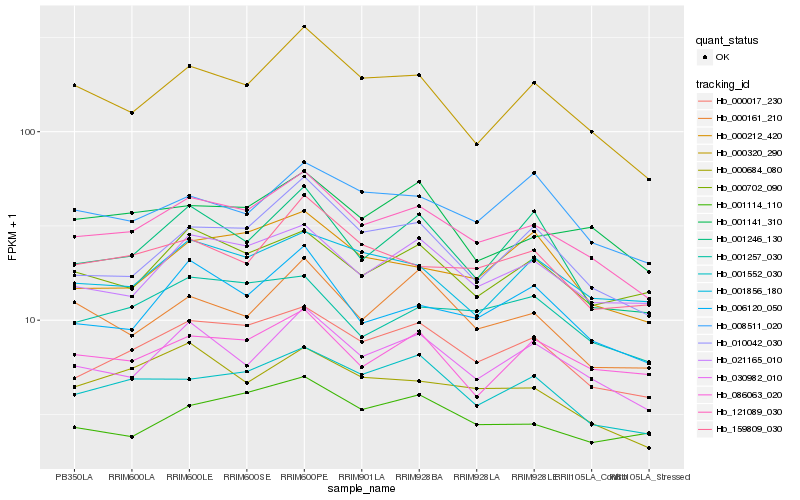
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_121089_030 |
0.0 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 2 |
Hb_000017_230 |
0.0542252688 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 3 |
Hb_030982_010 |
0.0616871474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_021165_010 |
0.0666907463 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000320_290 |
0.0676992739 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 6 |
Hb_001552_030 |
0.0690796157 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_010042_030 |
0.0759439355 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 8 |
Hb_006120_050 |
0.076818173 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 9 |
Hb_001141_310 |
0.0787302464 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 10 |
Hb_159809_030 |
0.0792485352 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_001257_030 |
0.0800866683 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek5 [Jatropha curcas] |
| 12 |
Hb_000702_090 |
0.0802221872 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 13 |
Hb_000684_080 |
0.0814503749 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 14 |
Hb_001856_180 |
0.0840193445 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 15 |
Hb_001114_110 |
0.0841497835 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 16 |
Hb_000212_420 |
0.084292899 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 17 |
Hb_000161_210 |
0.0853667248 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 18 |
Hb_001246_130 |
0.0854673913 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 19 |
Hb_008511_020 |
0.0865127082 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 20 |
Hb_086063_020 |
0.0867509958 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |