| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
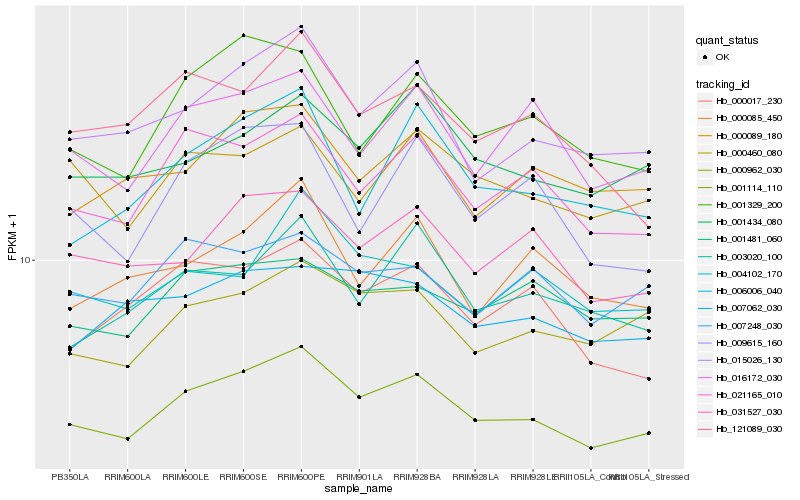
Hb_001114_110 |
0.0 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 2 |
Hb_015026_130 |
0.0609280973 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 3 |
Hb_021165_010 |
0.0618861698 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000962_030 |
0.0701907266 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 5 |
Hb_031527_030 |
0.07187997 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 6 |
Hb_016172_030 |
0.0747241834 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 7 |
Hb_001434_080 |
0.0761871829 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 8 |
Hb_001329_200 |
0.078163197 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000089_180 |
0.0785623922 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 10 |
Hb_003020_100 |
0.0807532977 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 11 |
Hb_000017_230 |
0.0811151625 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 12 |
Hb_006006_040 |
0.0812044383 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 13 |
Hb_007248_030 |
0.081543821 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 14 |
Hb_000460_080 |
0.0817558454 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |
| 15 |
Hb_001481_060 |
0.0819819853 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000085_450 |
0.0836131689 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 17 |
Hb_009615_160 |
0.0837524768 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 18 |
Hb_121089_030 |
0.0841497835 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 19 |
Hb_004102_170 |
0.0846107676 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 20 |
Hb_007062_030 |
0.0846222374 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |