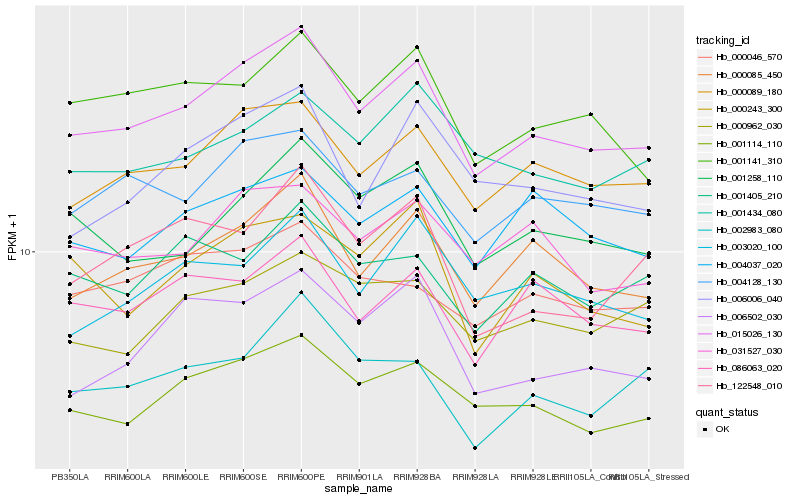
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015026_130 |
0.0 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 2 |
Hb_000089_180 |
0.0577157069 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 3 |
Hb_000085_450 |
0.0581300255 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 4 |
Hb_001114_110 |
0.0609280973 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 5 |
Hb_004128_130 |
0.0706632901 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001141_310 |
0.0738608267 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 7 |
Hb_001258_110 |
0.074975317 |
- |
- |
transporter, putative [Ricinus communis] |
| 8 |
Hb_031527_030 |
0.0768849857 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 9 |
Hb_086063_020 |
0.0795657799 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 10 |
Hb_003020_100 |
0.0795792419 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 11 |
Hb_006006_040 |
0.0807952872 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 12 |
Hb_000243_300 |
0.0832674152 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_000046_570 |
0.0835900964 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 14 |
Hb_006502_030 |
0.0841653869 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 15 |
Hb_004037_020 |
0.0849695239 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001434_080 |
0.085393375 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 17 |
Hb_122548_010 |
0.085887002 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 18 |
Hb_000962_030 |
0.0861880761 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 19 |
Hb_001405_210 |
0.0864822348 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 20 |
Hb_002983_080 |
0.0866371119 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |