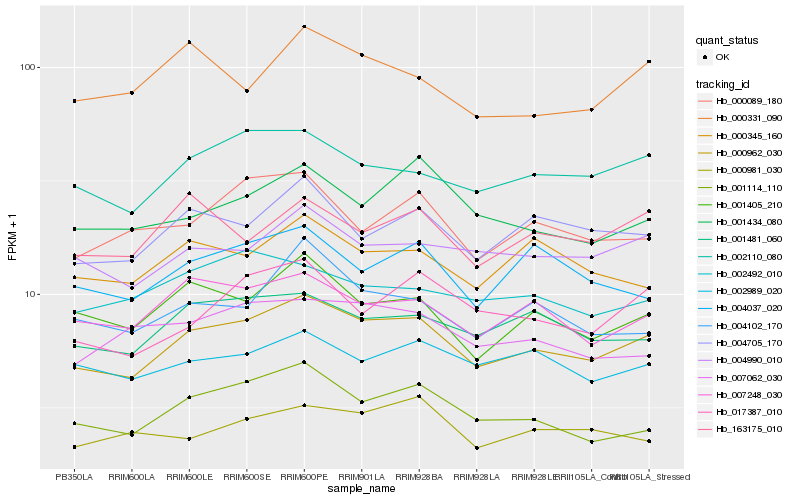
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000962_030 |
0.0 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 2 |
Hb_002110_080 |
0.0660760863 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 3 |
Hb_001114_110 |
0.0701907266 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 4 |
Hb_001481_060 |
0.0705994549 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007248_030 |
0.0710433298 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 6 |
Hb_004705_170 |
0.0724706562 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 7 |
Hb_001405_210 |
0.0738091562 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 8 |
Hb_004990_010 |
0.0742513732 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 [Jatropha curcas] |
| 9 |
Hb_017387_010 |
0.077374872 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 10 |
Hb_002989_020 |
0.0795850757 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001434_080 |
0.0799645237 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 12 |
Hb_000089_180 |
0.0810386833 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 13 |
Hb_004037_020 |
0.0823395035 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000345_160 |
0.082366337 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 15 |
Hb_007062_030 |
0.0824429127 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 16 |
Hb_004102_170 |
0.0825953616 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 17 |
Hb_163175_010 |
0.0838791242 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 18 |
Hb_002492_010 |
0.0856794264 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 19 |
Hb_000331_090 |
0.0858091755 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 20 |
Hb_000981_030 |
0.0858213622 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |